Abstract
The genus Themus Motschulsky, 1858 is one of the largest genera of beetle family Cantharidae, however, no complete mitochondrial genome is available in the public database so far. Here, we sequenced and analyzed the first complete mitochondrial genome of Themus, with T. foveicollis as the representative species. The mitochondrial genome was 16,469 bp in size with 37 genes including 13 protein-coding genes (PCGs), 22 transfer RNA genes, and two ribosomal RNA genes, as well as a non-coding region, which are arranged in the same way as the hypothetical ancestral insect. It exhibited a typical high A + T bias and a higher proportion of bases A and C than T and G (A: 41.1%, T: 37.5%, G: 8.6%, C: 12.9%). The phylogenetic trees of Elateroidea were reconstructed based on 13 PCGs sequences using the Bayesian inference (BI) and maximum-likelihood (ML) analyses to investigate its phylogenetic position. The results showed that Themus was consistently grouped with the remaining genera of Cantharidae and placed in a monophyletic clade of Cantharinae in high supporting values, which are congruent with the morphological classification. The newly sequenced mitochondrial genome in this study constitutes important reference data for reconstructing phylogeny of the family Cantharidae or the superfamily Elateroidea.
1. Introduction
Cantharidae, commonly known as soldier beetles, is occurring on all the world’s habitable continents (Delkeskamp Citation1977, Citation1978). The cantharid beetles spend the vast majority of their individual life spans as larvae occurring in micro-habitats with relative high humidity, such as beneath leaf litter, under stones, in loose soil, or under loose bark of decaying logs (Ramsdale Citation2010). The larvae prey on various invertebrates, including earth worms, slugs, and other insects (Balduf Citation1935; Janssen Citation1963; Langenstück et al. Citation1998), so they act on an important role in the ecosystem. Themus Motschulsky, 1858 is one of the largest genera of Cantharidae, with over 250 species widely distributed in the Palearctic and Oriental regions (Švihla Citation2008). To date, research on the genus Themus is limited to morphological classification (Yang et al. Citation2012, Citation2013). Although it has a high species diversity, no mitochondrial genome of Themus is available in the public database now (National Center for Biotechnology Information [NCBI], https://www.ncbi.nih.gov, accessed on 18 February 2023). The lack of genetic information limits our investigation in the phylogenetic relationship of this genus, thereby hindering our understanding of the evolution of Cantharidae or Elateroidea. Fortunately, we successfully sequenced and annotated the complete mitochondrial genome of Themus foveicollis (Fairmaire Citation1900) recently, and it is necessary for us to report it herein.
Themus foveicollis is a member restricted to eastern China (Fujian) (Kazantsev and Brancucci Citation2007). The adults are frequently met on the flowers or shrubs, sometimes can be attracted by lights in the evening. They are easily recognized by the large-sized body (ca. 13 mm in length), black antennae but yellow at basal antennomeres, green and metallic shining elytra, subquadrate, and yellow pronotum, as well as yellow legs except for black tarsi ().
Figure 1. The male habitus of Themus foveicollis in dorsal view. Scale bar: 2 mm. The picture was taken by Ya Kang.

In this study, the characteristics of the mitogenome of T. foveicollis were analyzed, and the phylogenetic trees were produced to investigate its phylogenetic position within Elateroidea. The mitogenome produced here will thus be an important reference data for reconstructing phylogeny of the family Cantharidae or the superfamily Elateroidea.
2. Materials
The material was collected in Fengyang Mountain (N:27°56′21″, E:119°12′58″; 2 May 2008) from Zhejiang Province. The sampling material was preserved in 100% ethanol at −20 °C. The specimen was deposited at the Museum of Hebei University, Baoding, China (MHBU) (http://museum.hbu.cn/, Yuxia Yang, [email protected]) under the voucher number 2-CA138.
3. Methods
3.1. Mitochondrial genome assembly and annotation
Total DNA was extracted from the thoracic muscle using a DNeasy Blood & Tissue kit. The mitogenome was sequenced using an Illumina Novaseq 6000 platform. High-quality reads were assembled using an iterative De Bruijn graph de novo assembler, the IDBA-UD toolkit (Peng et al. Citation2012). The gene COI was amplified through polymerase chain reaction using universal primers (Table S1) as ‘reference sequence’ to target mitochondrial scaffold. The assembled mitogenome sequence was annotated using Geneious version 2019.2 (Kearse et al. Citation2012) with the sequence of Lycocerus asperipennis (Wang et al. Citation2019) as a reference. Protein-coding genes (PCGs) were determined by identifying open reading frames, and the secondary structure and positions of 22 tRNAs were predicted using MITOS WebServer (http://mito.bioinf.uni-leipzig.de/index.py) (Bernt et al. Citation2013). Genomic sequence data was registered in the NCBI database under accession no. OQ621596. Mitogenomic circular map was drawn using the Draw Organelle Genome Maps (OGDRAW) (https://chlorobox.mpimp-golm.mpg.de/ OGDraw.html) (Lohse et al. Citation2007). The raw reads were mapped to the mitogenomic sequence using bwa version 0.7.17 (Li and Durbin Citation2009) and sorted using the samtools version 1.9 (Li et al. Citation2009), then the depth was calculated using bedtools version 2.27.1 (Maurano et al. Citation2012) and the figure was plotted by the barplot function in R version 3.4.3.
3.2. Phylogenetic analysis
A total of 24 species of the Elateroidea were chosen as ingroups, and another three species of Elateriformia were selected as outgroups (). The protein-coding sequences were aligned using MAFFT version 7.313 (Katoh and Standley Citation2013). After removing gaps and ambiguous sites using trimAI (Capella-Gutiérrez et al. Citation2009), we concatenated the sequences using PhyloSuite version 1.2.2 (Zhang et al. Citation2020). The optimal partition schemes and substitution models under codon mode were predicted using PartitionFinder2 (Lanfear et al. Citation2012) with the Bayesian information criterion (Tables S2 and S3).
Table 1. Information on the mitogenomes of the species used in this study.
Phylogenies were extrapolated by Bayesian inference (BI) and maximum-likelihood (ML) methods. The ML trees were constructed with ultrafast 5000 bootstrap replicates based on partition schemes of the genes and codons position using IQ-Tree version 1.6.8 (Nguyen et al. Citation2015). The BI tree was produced with four parallel runs for 10 million Markov Chain Monte Carlo (MCMC) generations based on partition schemes of codons position by MrBayes version 3.2.6 (Ronquist and Huelsenbeck Citation2003). Phylogenetic trees were visualized and annotated using the Interactive Tree of Life (ITOL) (https://itol.embl.de/itol.cgi) (Letunic and Bork Citation2021).
4. Results
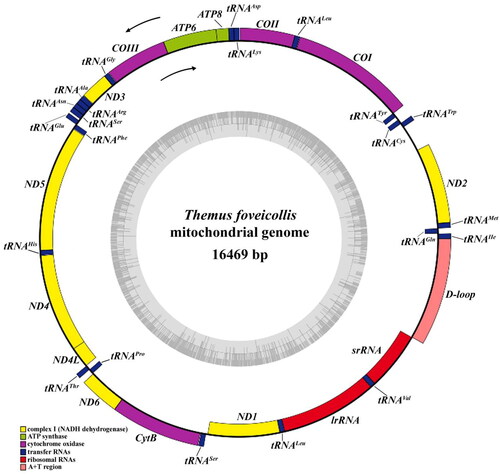
The complete mitochondrial genome of the Themus foveicollis was 16,469 bp in size with relatively high sequencing depth (Figure S1), which was composed of 14 genes (8 tRNAs, 4 PCGs, and 2 rRNAs) transcribed on the minority strand (N-strand), while the rest (14 tRNAs and 9PCGs) on the majority strand (J-strand) (). The composition of nucleotide bases for this mitogenome is as follows: A: 41.1%, T: 37.5%, G: 8.6%, C: 12.9%. All PCGs in mitogenome were initiated with the standard ATN codon and terminated with TAR (TAG/TAA) (Table S4).
Figure 2. Mitogenome map of Themus foveicollis sequenced in this study. Different colors indicate different types of genes and regions. Genes outside the black circle are located on the J-strand in counterclockwise, and those inside the black circle are located at the N-strand in clockwise.
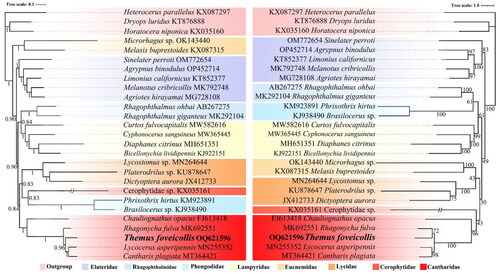
The topologies constructed by ML and BI analyses based on the PCGs dataset were slightly different ( and ). But T. foveicollis was consistently grouped with the remaining genera of Cantharidae (represented by Cantharis, Lycocerus, Rhagonycha, and Chauliognathus), which was robust and well-supported (BS = 96/95, PP = 1). Further Chauliognathus (as the sole representative of Chauliognathinae) was recovered sister to the others with high supporting values (BS = 96/95, PP = 1), which all belonged to the subfamily Cantharinae. However, within Cantharinae, Themus was recovered sister either to all others of Cantharinae (BS = 51, PP = 0.96) or to Rhagonycha with relatively low support values (BS = 72).
5. Discussion and conclusion
In this study, we successfully sequenced and annotated the first complete mitochondrial genome of the cantharid genus Themus, with T. foveicollis as the representative species. The newly sequenced mitogenome contains 37 genes and a non-coding region, which are arranged in the same way as the hypothetical ancestral insect (Clary and Wolstenholme Citation1985), and showing a typical high A + T bias (A + T: 78.6%, G + C: 21.4%) and a higher proportion of bases A and C than T and G, as other insects (Raupach et al. Citation2022). The inferred phylogeny indicated that Themus was consistently grouped with the remaining genera of Cantharidae and placed in a monophyletic clade of Cantharinae in high supporting values, which are congruent with the morphological classification (Kazantsev and Branucci 2007). However, within Cantharinae, the phylogenetic relationship between Themus and others remain unclear due to inconsistent topologies by different cladistic methods, which require more sampling data to clarify this aspect in future. Nevertheless, the newly sequenced mitochondrial genome in this study will facilitate further investigations into the phylogenetic relationship of the Cantharinae or Cantharidae, even Elateroidea.
Ethical approval
No specific permits were required for the insect specimens collected for this study. The field studies did not involve endangered or protected species. The insect species sequenced is a common Cantharidae species in China and is not included in the ‘List of Protected Animals in China’.
Author contributions
Conceptualization: Xiaoxiao Wang, Ya Kang, Guanglin Xie, and Yuxia Yang; methodology: Xiaoxiao Wang and Ya Kang; formal analysis: Xiaoxiao Wang and Ya Kang; investigation: Ya Kang, Yuxia Yang; writing – original draft preparation: Xiaoxiao Wang, Ya Kang, Yuxia Yang; writing – review and editing: Xiaoxiao Wang, Ya Kang, and Yuxia Yang; All authors have read and agreed to the published version of the manuscript.
Supplemental Material
Download MS Word (20.5 KB)Supplemental Material
Download MS Word (244 KB)Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The genome sequence data that support the finding of this study is openly available in GenBank of NCBI at https://www.ncbi.nih.gov under the accession no. OQ621596. The associated BioProject, SRA, and BioSample numbers are PRJNA941014, SRR23926838, and SAMN33775840, respectively.
Additional information
Funding
References
- Amaral DT, Mitani Y, Ohmiya Y, Viviani VR. 2016. Organization and comparative analysis of the mitochondrial genomes of bioluminescent Elateroidea (Coleoptera: polyphaga). Gene. 586(2):254–C2. doi:10.1016/j.gene.2016.04.009.
- Balduf WV. 1935. The bionomics of entomophagous coleoptera. New York (NY): J. S. Swift; p. 220.
- Bernt M, Donath A, Jühling F, Externbrink F, Florentz C, Fritzsch G, Pütz J, Middendorf M, Stadler PF. 2013. MITOS: improved de novo metazoan mitochondrial genome annotation. Mol Phylogenet Evol. 69(2):313–319. doi:10.1016/j.ympev.2012.08.023.
- Capella-Gutiérrez S, Silla-Martínez JM, Gabaldón T. 2009. TrimAI: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics. 25(15):1972–1973. doi:10.1093/bioinformatics/btp348.
- Chen X, Dong Z, Liu G, He J, Zhao R, Wang W, Peng Y, Li X. 2019. Phylogenetic analysis provides insights into the evolution of Asian fireflies and adult bioluminescence. Mol Phylogenet Evol. 140:106600. doi:10.1016/j.ympev.2019.106600.
- Clary DO, Wolstenholme DR. 1985. The mitochondrial DNA molecule of Drosophila yakuba: nucleotide sequence, gene organization, and genetic code. J Mol Evol. 22(3):252–271. doi:10.1007/BF02099755.
- Delkeskamp K. 1977. Cantharidae. In: Wilcox JA, editor. Coleopterorum catalogus supplementa, Pars 165, Fasc. 1. Vol. 2. Holland: W. Junk, Hague; p. 1–485.
- Delkeskamp K. 1978. Cantharidae. Corrigenda et addenda. In: Wilcox JA, editor. Coleopterorum catalogus supplementa, Pars 165, Fasc. 2. Vol. 2. Holland: W. Junk, Hague; p. 487–556.
- Fairmaire L. 1900. Description de Coléoptères nouveaux recueillis en Chine par M. de Latouche. Ann Soc Entomol France. 68(1899):616–649.
- Gerritsen AT, New DD, Robison BD, Rashed A, Hohenlohe P, Forney L, Rashidi M, Wilson CM, Settles ML. 2016. Full mitochondrial genome sequence of the sugar beet wireworm Limonius californicus (Coleoptera: Elateridae), a common agricultural pest. Genome Announc. 4(1):e01628–e01615. doi:10.1128/genomeA.01628-15.
- Janssen W. 1963. Untersuchungen zur morphologie, biologie und okologie von cantharis L. und Rhagonycha Eschsch (Cantharidae, Col). Zeitschrift Wissenschaftliche Zool. 169:115–202.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780. doi:10.1093/molbev/mst010.
- Kazantsev SV, Brancucci M. 2007. Cantharidae. In Löbl I, Smetana A, editor. Catalogue of palaearctic coleoptera. Vol. 4. Stenstrup, Denmark: Apollo Books; p. 234–298.
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, et al. 2012. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 28(12):1647–1649. doi:10.1093/bioinformatics/bts199.
- Lanfear R, Calcott B, Ho SY, Guindon S. 2012. PartitionFinder: combined selection of partitioning schemes and substitution models for phylogenetic analyses. Mol Biol Evol. 29(6):1695–1701. doi:10.1093/molbev/mss020.
- Langenstück C, Heimbach U, Larink O. 1998. Larven der Cantharidae (Insecta: coleoptera) auf Ackerflächen in So-Niedersachsen und Aspekte ihrer Biologie. Braunschweiger Naturkundliche Schriften. 5:551–568.
- Letunic I, Bork P. 2021. Interactive Tree Of Life (iTOL) v5: an online tool for phylogenetic tree display and annotation. Nucleic Acids Res. 49(W1):W293–W296. doi:10.1093/nar/gkab301.
- Li H, Durbin R. 2009. Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics. 25(14):1754–1760. doi:10.1093/bioinformatics/btp324.
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R, 2009. The sequence alignment/map format and SAMtools. Bioinformatics. 25(16):2078–2079. doi:10.1093/bioinformatics/btp352.
- Li W, Liu Q, Fu X. 2022. The complete mitochondrial genome of the firefly Curtos fulvocapitalis (Coleoptera: Lampyridae). Mitochondrial DNA B Resour. 7(1):1–3. doi:10.1080/23802359.2021.1958080.
- Linard B, Arribas P, Andújar C, Crampton-Platt A, Vogler AP. 2016. Lessons from genome skimming of arthropod-preserving ethanol. Mol Ecol Resour. 16(6):1365–1377. doi:10.1111/1755-0998.12539.
- Lin A, Zhao X, Song N, Zhao T. 2018. Analysis of the complete mitochondrial genome of click beetle Agriotes hirayamai (Coleoptera: Elateridae). Mitochondrial DNA B Resour. 3(1):290–291. doi:10.1080/23802359.2018.1443041.
- Li X, Ogoh K, Ohba N, Liang X, Ohmiya Y. 2007. Mitochondrial genomes of two luminous beetles, Rhagophthalmus lufengensis and R. ohbai (Arthropoda, Insecta, Coleoptera). Gene. 392(1–2):196–205. doi:10.1016/j.gene.2006.12.017.
- Liu HY, Kang ZX, Zhang F, Ge XY, Yang YX. 2019. The complete mitogenome of Lycostomus sp. (Elateroidea: Lycidae). Mitochondrial DNA B Resour. 4(2):3813–3815. doi:10.1080/23802359.2019.1682483.
- Lohse M, Drechsel O, Bock R. 2007. Organellar Genome DRAW (OGDRAW): a tool for the easy generation of high-quality custom graphical maps of plastid and mitochondrial genomes. Curr Genet. 52(5–6):267–274. doi:10.1007/s00294-007-0161-y.
- Maurano MT, Humbert R, Rynes E, Thurman RE, Haugen E, Wang H, Reynolds AP, Sandstrom R, Qu H, Brody J, et al. 2012. Systematic localization of common disease-associated variation in regulatory DNA. Science. 337(6099):1190–1195. doi:10.1126/science.1222794.
- Nguyen LT, Schmidt HA, Von Haeseler A, Minh BQ. 2015. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 32(1):268–274. doi:10.1093/molbev/msu300.
- Peng Y, Leung HC, Yiu SM, Chin, FY, 11. 2012. IDBA-UD: a de novo assembler for single-cell and metagenomic sequencing data with highly uneven depth. Bioinformatics. 28(11):1420–1428. doi:10.1093/bioinformatics/bts174.
- Ramsdale AS. 2010. Cantharidae Imhoff, 1856. In: Leschen RAB, Beutel RG, Lawrence JF, editors. Coleoptera, beetles, handbuch der zoologie [Handbook of zoology], Vol. 2. Berlin, Germany: Walter de Gruyter Inc.; p. 153–162.
- Raupach MJ, Deister F, Villastrigo A, Balke M. 2022. The complete mitochondrial genomes of Notiophilus quadripunctatus Dejean, 1826 and Omophron limbatum (Fabricius, 1777): new insights into the mitogenome phylogeny of the Carabidae (Insecta, Coleoptera). Insect Syst Evol. 53(3):242–263. doi:10.1163/1876312X-bja10027.
- Ronquist F, Huelsenbeck JP. 2003. MrBayes 3: bayesian phylogenetic inference under mixed models. Bioinformatics. 19(12):1572–1574. doi:10.1093/bioinformatics/btg180.
- Sheffield NC, Song H, Cameron SL, Whiting MF. 2009. Nonstationary evolution and compositional heterogeneity in beetle mitochondrial phylogenomics. Syst Biol. 58(4):381–394. doi:10.1093/sysbio/syp037.
- Švihla V. 2008. Redescription of the subgenera of the genus Themus Motschulsky, 1858, with description of five new species (Coleoptera: cantharidae). Veröffentlichungen Naturkundemuseums Erfurt. 27:183–190.
- Uribe JE, Gutiérrez-Rodríguez J. 2016. The complete mitogenome of the trilobite beetle, Platerodrilus sp. (Elateroidea: Lycidae). Mitochondrial DNA B Resour. 1(1):658–659. doi:10.1080/23802359.2016.1219626.
- Wang Y, Liu Y. 2019. The complete mitochondrial genome of Melanotus cribricollis (Coleoptera: Elateridae) and phylogenetic analysis. Mitochondrial DNA B Resour. 4(2):3238–3239. doi:10.1080/23802359.2019.1669085.
- Wang P, Yuan LL, Ge XY, Liu HY, Yang YX. 2019. The complete mitochondrial genome sequence and phylogenetic analysis of Lycocerus asperipennis (Coleoptera: cantharidae). Mitochondrial DNA B Resour. 4(2):3768–3769. doi:10.1080/23802359.2019.1682478.
- Xi HC, Ge SJ, Kang ZX, Liu HY, Yang YX. 2020. The complete mitochondrial genome sequence and phylogenetic analysis of Cantharis plagiata (Coleoptera, Canthridae). Mitochondrial DNA B Resour. 5(3):2386–2388. doi:10.1080/23802359.2020.1775515.
- Yang Z, Fu X. 2019. The complete mitochondrial genome of the firefly, Diaphanes citrinus (Olivier), (Coleoptera: Lampyridae). Mitochondrial DNA B Resour. 4(2):2986–2987. doi:10.1080/23802359.2019.1664351.
- Yang Y, Kopetz A, Yang X. 2013. Taxonomic and nomenclatural notes on the genera Themus Motschulsky and Lycocerus Gorham (Coleoptera, Cantharidae). Zookeys. 340(340):1–19. doi:10.3897/zookeys.340.5470.
- Yang YX, Lu YY, Yang XK. 2012. Four newly recorded to China species of the genus Themus Motschulsky, 1857 (Coleoptera: Cantharidae). Far Eastern Entomol. 242:1–6.
- Yuan L, Ge X, Xie G, Liu H, Yang YX. 2021. First Complete mitochondrial genome of Melyridae (Coleoptera, Cleroidea): genome description and phylogenetic implications. Insects. 12(2):87. doi:10.3390/insects12020087.
- Zhang D, Gao F, Jakovlić I, Zou H, Zhang J, Li WX, Wang GT. 2020. PhyloSuite: an integrated and scalable desktop platform for streamlined molecular sequence data management and evolutionary phylogenetics studies. Mol Ecol Resour. 20(1):348–355. doi:10.1111/1755-0998.13096.