Abstract
Pleuronectidae is a well-studied familyin the order Pleuronectiformes. In contrast, genetic research on the flatfish Acanthopsetta nadeshnyi of the Pleuronectidae family is limited. This study reports the complete mitogenome of A. nadeshnyi. The mitogenome was 17,206 bases long and included 13 protein-coding genes (PCGs), 22 transfer RNA (tRNA) genes, two ribosomal RNA (rRNA) genes, and a putative control region. Phylogenetic analysis based on the nucleotide sequences of the 13 PCGs confirmed that A. nadeshnyi belongs to the Pleuronectidae family.
Introduction
The family Pleuronectidae (order Pleuronectiformes) is well-known. Several studies have revealed 23 genera and 61 species in the faily (Nelson et al. Citation2016). Traditionally, all known flatfish were assigned to the family Pleuronectidae (Jordan and Goss Citation1889), but were later divided into five families (Norman Citation1934). Acanthopsetta nadeshnyi (Schmidt 1904) is a Pleuronectidae flatfish that lives at depths from 18 to 900 m. It occurs from the Bering Sea and the Sea of Okhotsk to the waters of Japan and Korea (Masuda et al. Citation1984; Matarese et al. Citation1987).
The mitochondrial genome is often used to study the phylogenetic relationships between diverse species because of its short length and maternal inheritance (Miya et al. Citation2003; Pardo et al. Citation2005). In addition, its gene structure, content, and sequence are conserved among vertebrates. Despite the value of mitochondrial DNA (mtDNA) in identifying similarities between organisms, there are only a few studies on A. nadeshnyi; Genbank contains only 13 sequences, including seven mitochondrial sequences of two cox1, two cytb, one 12S rRNA, and one trnP. Here, we determined the complete mitochondrial genome of A. nadeshnyi and determinedits phylogenetic position ofaccordingly. This mitogenome extends the available genetic data on A. nadeshnyi and enables the determination of its phylogenetic position.
Materials
A specimen was provided by the Marine Bio-Resource Information System (MBRIS; Voucher No. MRS002000103676; https://www.mbris.kr/pub/main/publicMainPage.do; [email protected]). Specimen was collected from Goseong, South Korea (34°90 ′N, 128°22 ′E) and deposited at the Pukyong National University storage facility ().
Methods
Genomic DNA was purified using a PureHelix™ Genomic DNA Prep Kit [Animals], Solution Type (NANOHELIX, Daejeon), and the cox1 gene was amplified using a fish universal primer based on Ward et al. (Citation2005). The amplicon sequence was analyzed by Macrogen (Seoul), and BLASTN searches against every nucleotide sequence in GenBank (Johnson et al. Citation2008) was conducted to identify the sample species. In addition, p-distance analysis was performed using Mega software version 11 (Tamura et al. Citation2021) with 1000 bootstrap replicates using the three most similar cox1 sequences from the BLASTN results.
Library preparation was performed with an MGIEasy DNA Library Prep Set (MGI, Shenzhen) according to the manufacturer’s instructions. Raw data were generated by next-generation sequencing (NGS) using an MGISEQ-2000 platform, and deposited in the SRA database (Accession No. SRR22019788). The adapter was trimmed using Cutadapt version 4.1 (Martin Citation2011), and contig assembly was performed using CLC Genomics Workbench version 20.04 (QIAGEN, Hilden) using the default options in the de novo assembler. A circular sequence was mapped with the trimmed data sets using Geneious Software version 20.2.2 (https://www.geneious.com) to verify the complete mtDNA sequence of A. nadeshnyi. Tandem repeats and direct repeat motifs were predicted using Geneious software. Annotation was conducted on the MITOS WebServer (Bernt et al. Citation2013) and manually corrected using SnapGene software version 5.3.2 (GSL Biotech LLC, https://snapgene.com). The final mtDNA sequence was registered in GenBank (Accession No. OP028121). A mitogenome map was prepared using the CGView Server (Grant and Stothard Citation2008).
A maximum likelihood (ML) phylogenetic tree was constructed using MEGA software version 11 (Tamura et al. Citation2021) using mitogenomes from all 18 members of the family Pleuronectidae in GenBank and one member of the family Acipenseridae, Acipenser dabryanus (Accession No. AY510085), was used as the outgroup. All mitogenome sequences () were collected from GenBank. Nucleotide sequences of protein-coding genes (PCGs) were gathered from each mitogenome sequence and aligned using the ClustalW multiple alignment tool in BioEdit with default options. Phylogenetic analysis was conducted using the GTR + G + I model with 1000 bootstrap replicates.
Table 1. Mitogenome sequence used in this study.
Results
BLASTN of cox1 obtained by PCR showed an identity match of 99.85% with that of A. nadeshnyi (Accession No. MH032397), 95.98% with that of Dexistes rikuzenius (Accession No. MH032411), and 95.09% with that of Hippoglossoides elassodon (Accession No. JQ354126). The p-distance were 1.35 (A. nadeshnyi), 5.29 (D. rikuzenius), and 5.84 (H. elassodon), reflecting the same pattern as the BLASTN results. These results confirm that our flatfish specimen is A. nadeshnyi.
In total, 20,481,617 read-pairs were produced by NGS, from which the mitogenome of A. nadeshnyi (17,206 bp) was assembled with a mean coverage of 513.0 ± 97.9 (Figure S1). The mitogenome encoded 37 genes, including 13 PCGs, 22 transfer RNA (tRNA) genes, and two ribosomal RNA (rRNA) genes (). Every PCG was encoded on the positive strand except nad6, which was encoded on the negative strand. ATG was a start codon in 12 PCGs (nad1, nad2, cox2, atp8, atp6, cox3, nad3, nad4l, nad4, nad5, nad6, and cob), whereas only cox1 started with the GTG codon. Transcription was stoped with the TAA codon in six genes (nad1, atp8, atp6, nad4l, and nad5, nad6). In contrast, nad2, cox2, nad3, nad4, and cob stoped with truncated T-, and cox3 terminated with truncated TA-.
Figure 2. Complete mitochondrial genome map of Acanthopsetta nadeshnyi. The map comprised 37 genes, including 13 protein-coding genes, 22 transfer RNA genes, and two ribosomal RNA genes. The arrowheads indicate the direction of transcription. Highly similar 58–74 bp direct repeat motifs occur in the putative control region (black box).
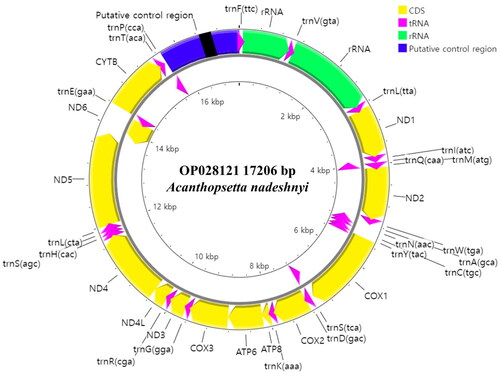
Including two trnL and two trnS, the mitogenome of A. nadeshnyi comprised 22 tRNA genes 14 tRNA genes were encoded on the positive strand and the others were encoded on the negative strand ().
The small rRNA gene was 949 bases long and located between trnF and trnV. The large rRNA was 1714 bases long and is located between trnV and trnL(tta). The putative control region was 1528 bases long and located between trnP and trnF. Conserved sequence box D (5′- CCTGGCATTTGGTTCC-3′), a pyrimidine sequence run (5′- TTCTCTTTTTTTTTTTCCTTTC-3′), and two conserved sequence boxes (CSB-2: 5′-AAACCCCCCTACCCCCC-3′; CSB-3: 5′-TGAAAACCCCCCGGAAACA-3′) occurred in the putative control region. There were no tandem repeats, but highly similar 58–74 bp direct repeat motifs were found at positions 802–1152 of the putative control region. In addition, the OL region was observed in another non-coding region that may fold into a stem-loop secondary structure.
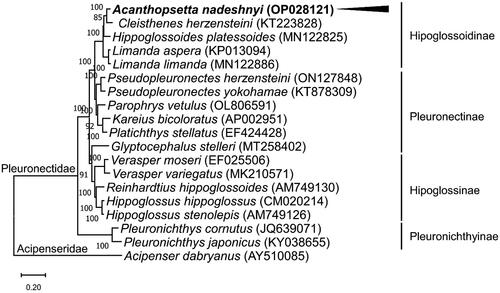
The ML phylogenetic tree of the family Pleuronectidae showed that A. nadeshnyi was grouped with Cleisthenes herzensteini (). The mitogenome of A. nadeshnyi was separated into different nodes, including species of the subfamily Hippoglossoidinae.
Figure 3. Phylogenetic tree of Acanthopsetta nadeshnyi and related species. Based on the maximum likelihood (ML) tree, the phylogenetic position of A. nadeshnyi was analyzed using 13 PCGs from the mitogenomes of 18 members of the Pleuronectidae family with Acipenser dabryanus (Accession No. AY510085) as an outgroup. GenBank accession numbers for the mitogenome sequences are provided next to the species names. The node numbers correspond to the posterior probabilities of Bayesian inference. The black arrow and bold fonts highlight the mitogenome of A. nadeshnyi.
Discussion and conclusion
Here, we reported that the complete mtDNA sequence of A. nadeshnyi had a comparable gene composition with that generally observed in vertebrate mtDNA (Kolesnikov and Gerasimov Citation2012). The composition and gene order of this mitogenome matched those of teleosts, reflecting the limited occurrence of mitogenome rearrangements in fish (Gong et al. Citation2013). In the Pleuronectidae family, various tandem repeats contribute to the relatively large size of the control regions (Zheng et al. Citation2016; Song et al. Citation2017). However, no tandem repeats were found in the mitogenome of A. nadeshnyi similar to that for G. stelleri (Kim and Jang Citation2022), whereas highly similar direct repeat motifs and several conventional motifs were observed. These highly similar direct repeat motifs can potentially extend the size of the control region. This suggests that tandem repeats and highly similar direct repeat motifs may be useful biomarkers for identifying relationships between Pleuronectidae species. Considering the limited number of studies on A. nadeshnyi, the present study contributes new genetic information for Pleuronectidae.
Ethical approval
No ethical approval is required for this study. We used a flatfish fin from a specimen that was previously collected by the MBRIS, outside of this study. This specimen was dead, and the sample was provided with permission by the MBRIS (permission no. 2022-130).
Authors’ contributions
JYC, MSK, JKK, and HHL conceived the original idea. JYC conducted the experiments. JYC wrote the manuscript with support from MSK and HHL. TWK and JHK performed data analysis, and JKK supplied specimen. All authors agree to be accountable for all aspects of the work.
Supplemental Material
Download MS Word (70.1 KB)Supplemental Material
Download PDF (297.1 KB)Acknowledgment
Dr. Khawaja Muhammad Imran Bashir reviewed this article for the English language; the authors are thankful for his suggestions and support.
Disclosure statement of interest
No potential conflict of interest was reported by the author(s).
Data availability statement
The genome sequence data supporting this study’s findings are available in GenBank of NCBI at (https://www.ncbi.nlm.nih.gov/) under accession no. OP028121. The associated BioProject, SRA, and Bio-Sample numbers are PRJNA862354, SRR22019788, and SAMN29974750, respectively.
Additional information
Funding
References
- Bernt M, Donath A, Jühling F, Externbrink F, Florentz C, Fritzsch G, Pütz J, Middendorf M, Stadler PF. 2013. MITOS: improved de novo metazoan mitochondrial genome annotation. Mol Phylogenet Evol. 69(2):313–319. doi: 10.1016/j.ympev.2012.08.023.
- Bo Z, Wenping S, Kefeng L, Debin Z, Chao M, Guangxia X. 2016. The complete mitochondrial genome of Cleithenes herzenstein and its phylogenetic analysis. Mitochondrial DNA Part A DNA Mapp Seq Anal. 27(5):3663–3665. doi: 10.3109/19401736.2015.1079845.
- Chae JY, Kim JH, Kang TW, Kim DG, Lee HH, Kim MS. 2022. The complete mitochondrial genome of Pseudopleuronectes herzensteini. Mitochondrial DNA Part B Resour. 7(7):1305–1307. doi: 10.1080/23802359.2022.2095234.
- Gong L, Shi W, Si LZ, Kong XY. 2013. Rearrangement of mitochondrial genome in fishes. Zool Res. 34(6):666–673.
- Grant JR, Stothard P. 2008. The CGView Server: a comparative genomics tool for circular genomes. Nucleic Acids Res. 36(Web Server issue):W181–W184. doi: 10.1093/nar/gkn179.
- He C, Han J, Ge L, Zhou Z, Gao X, Mu Y, Liu W, Cao J, Liu Z, He C, et al. 2008. Sequence and organization of the complete mitochondrial genomes of spotted halibut (Verasper variegatus) and barfin flounder (Verasper moseri). DNA Seq. 19(3):246–255. doi: 10.1080/10425170701563303.
- Johnson M, Zaretskaya I, Raytselis Y, Merezhuk Y, McGinnis S, Madden TL. 2008. NCBI BLAST: a better web interface. Nucleic Acids Res. 36(Web Server issue):W5–W9. doi: 10.1093/nar/gkn201.
- Jordan DS, Goss DK. 1889. A review of the flounders and soles (Pleuronectidae) of America and Europe.
- Kim YJ, Jang YS. 2022. Comparative analysis of mitochondrial genomes of Pleuronectidae (Pleuronectiformes) in the seas around the Korean Peninsula. KOFFST International Conference 2022. p. 242–242.
- Kolesnikov AA, Gerasimov ES. 2012. Diversity of mitochondrial genome organization. Biochemistry. 77(13):1424–1435. doi: 10.1134/S0006297912130020.
- Lim HK, Jung HS, Yoon M, Lee SH, Kim DS. 2019. Complete mitochondrial genomes of two Pleuronectid species: Clidoderma asperrimum and Verasper variegatus (Teleostei: Pleuronectiformes: Pleuronectidae). Mitochondrial DNA Part B Resour. 4(2):3931–3932. doi: 10.1080/23802359.2019.1687343.
- Liu J, Tan C, Xiao K, Wang B, Liu X, Du H. 2017. The complete mitochondrial genome of Dabry’s sturgeon (Acipenser dabryanus). Mitochondrial DNA Part B Resour. 2(1):54–55. doi: 10.1080/23802359.2017.1280698.
- Martin M. 2011. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet j. 17(1):10–12. doi: 10.14806/ej.17.1.200.
- Masuda H, Amaoka K, Araga C, Uyeno T, Yoshino T. 1984. The Fishes of the Japanese Archipelago. 1
- Matarese AC, Kendall AW, Jr Blood DM, Vinter BM. 1987. Laboratory guide to early life history stages of Northeast Pacific Fishes. Laguna. 53:56.
- Miya M, Kawaguchi A, Nishida M. 2001. Mitogenomic exploration of higher teleostean phylogenies: a case study for moderate-scale evolutionary genomics with 38 newly determined complete mitochondrial DNA sequences. Mol Biol Evol. 18(11):1993–2009. doi: 10.1093/oxfordjournals.molbev.a003741.
- Miya M, Takeshima H, Endo H, Ishiguro NB, Inoue JG, Mukai T, Satoh TP, Yamaguchi M, Kawaguchi A, Mabuchi K, et al. 2003. Major patterns of higher teleostean phylogenies: a new perspective based on 100 complete mitochondrial DNA sequences. Mol Phylogenet Evol. 26(1):121–138. doi: 10.1016/s1055-7903(02)00332-9.
- Mjelle KA, Karlsen BO, Jørgensen TE, Moum T, Johansen SD. 2008. Halibut mitochondrial genomes contain extensive heteroplasmic tandem repeat arrays involved in DNA recombination. BMC Genomics. 9(1):10. doi: 10.1186/1471-2164-9-10.
- Nelson JS, Grande TC, Wilson MV. 2016. Fishes of the World. New York: John Wiley & Sons.
- Norman JR. 1934. A systematic monograph of the flatfishes (Heterosomata). London: Bulletin of the British Museum (Natural History).
- Pardo BG, Machordom A, Foresti F, Porto-Foresti F, Azevedo MF, Bañon R, Sánchez L, Martínez P. 2005. Phylogenetic analysis of flatfish (Order Pleuronectiformes) based on mitochondrial 16s rDNA sequences. Sci Mar. 69(4):531–543. doi: 10.3989/scimar.2005.69n4531.
- Shi W, Dong XL, Wang ZM, Miao XG, Wang SY, Kong XY. 2013. Complete mitogenome sequences of four flatfishes (Pleuronectiformes) reveal a novel gene arrangement of L-strand coding genes. BMC Ecol Evol. 13(1):1–9.
- Song Y, Song N, Zhou Y, Li P, Zhao S, Gao T. 2017. Complete mitochondrial genome of the Pleuronichthys lighti (Pleuronectiformes, Pleuronectidae) with phylogenetic consideration. Genes Genom. 39(11):1271–1284. doi: 10.1007/s13258-017-0570-3.
- Tamura K, Stecher G, Kumar S. 2021. MEGA11: molecular evolutionary genetics analysis version 11. Mol Biol Evol. 38(7):3022–3027. doi: 10.1093/molbev/msab120.
- Ward RD, Zemlak TS, Innes BH, Last PR, Hebert PD. 2005. DNA barcoding Australia’s fish species. Philos Trans R Soc Lond B Biol Sci. 360(1462):1847–1857. doi: 10.1098/rstb.2005.1716.
- Zheng F, Liu H, Zhang Y, Wang Q, Wang B. 2016. Complete mitochondrial genome of the Pseudopleuronectes yokohamae (Pleuronectiforll: pleuronectidae). Mitochondrial DNA Part A DNA Mapp Seq Anal. 27(6):4347–4348. doi: 10.3109/19401736.2015.1089494.

