Abstract
Spurilla braziliana MacFarland 1909 is a morphologically diverse nudibranch found in the Pacific and Western Atlantic. The complete mitochondrial genome of S. braziliana has been constructed using next-generation sequencing technology. The mitochondrial genome is 14,291 bp and contains 13 protein-coding genes, 2 rRNA genes, and 23 tRNA genes. Molecular phylogenetic analysis using the maximum likelihood method revealed that S. braziliana is included in the superfamily Aeolidioidea and forms a monophyletic group with Berghia stephanieae, a nudibranch of the family Aeolidiidae. This study reinforces existing taxonomic insights and provides a basis for further molecular phylogenetic analysis.
Introduction
Members of the genus Spurilla are unique nudibranchs capable of storing and utilizing cnidarian nematocysts in their bodies (Greenwood and Mariscal Citation1984). These sea slugs are widely distributed in the world’s oceans and attract attention from a taxonomic perspective due to their diverse coloration and morphological characteristics (Carmona et al. Citation2014). Previous research by Carmona et al. (Carmona et al. Citation2013) has shown that only one species of Spurilla, S. braziliana, is found in the Pacific Ocean. However, there is significant morphological diversity within S. braziliana (Carmona et al. Citation2014) suggesting that further molecular phylogenetic analysis may be required to distinguish the different morphotypes. This study reports the complete mitochondrial genome sequence of one morphotype of S. braziliana from Japan.
Materials
A sample of Spurilla braziliana was collected on 10 June 2021 from the intertidal area of Miura City (35°16′18.8′′N 139°34′04.7′′E), Kanagawa Prefecture, Japan () and preserved in NucleoProtect RNA (MACHEREY-NAGEL) for 5.5 months at −80 °C before DNA extraction. The sample was then transferred to 99% ethanol for deposition at the University Museum of the University of Tokyo (http://www.um.u-tokyo.ac.jp/, Assoc. Prof. Takenori Sasaki, [email protected]) under voucher number RM34044.
Figure 1. The individual of S. braziliana used in this study. Its morphotype is similar to the one shown in Figure 4(E) in the previous taxonomic study (Carmona et al. Citation2014). The picture was taken by Hideaki Mizobata.

Methods
Prior to the sequencing process, genomic DNA was extracted from 25 mg of the body wall of the aforementioned sample using the DNeasy Blood & Tissue Kit (QIAGEN, Hilden, Germany) according to the manufacturer’s instructions. A DNAlibrary for PE150 sequencing was prepared with MGIEasy FS DNA Library Prep Set (MGI) and sequenced with DNBSEQ-T7 (MGI) at Genome-Lead Corporation, Japan. A total of 63.6 Gbp of sequence data was obtained from the sequencing, with a Q30 score of 94.08%, indicating sufficient quality for assembly. The reads were assembled with CLC Genomics Workbench version 8.5 (QIAGEN Aarhus A/S) to construct a primary assembly. The primary assembly was polished using bwa mem version 0.7.15-r1140 (Li Citation2013), samtools version 1.4 (Danecek et al. Citation2021), and pilon version 1.22 (Walker et al. Citation2014) with the following commands.
$ bwa index assembly.fasta
$ bwa mem assembly.fasta read1.fastq read2.fastq|samtools view -bS|samtools sort > assembly.fasta.bam
$ samtools index assembly.fasta.bam
$ pilon –genome assembly.fasta –frags assembly.fasta.bam –outdir.
To further improve the polished primary assembly, the sequence was circularized by the ends, cut at a site differing from the circularization point, and polished once more. The raw reads were mapped to the final mitochondrial genome using bwa mem and samtools to ensure proper assembly (Supplementary Figure 1). The average coverage was confirmed to be 7866, further validating the robustness of the assembly and polishing. The polished mitochondrial genome was annotated with MITOS (Donath et al. Citation2019) and manually corrected using Geneious Prime Java version 11.0.14.1 + 1 (Biomatters Ltd., Auckland, New Zealand). MitoZ (Meng et al. Citation2019) was used for the accurate annotation of tRNA. The identified tRNA genes were initially folded using the RNA-fold web server (Gruber et al. Citation2008) to generate a rough secondary structure, which was then manually adjusted using VARNA (Darty et al. Citation2009). Annotations and other mitochondrial genome structures were visualized using OGDRAW (Greiner et al. Citation2019).
To confirm the species identity of our sample as S. braziliana, we performed nucleotide BLAST (blastn) analyses using the web interface of the National Center for Biotechnology Information (NCBI). The COX1 and 16S rRNA gene sequences from our specimen were used as queries, with the non-redundant nucleotide collection (nt) database chosen for the search. Default parameters were used for all other settings.
The 13 protein-coding genes within the mitochondrial genome were each individually aligned with their corresponding genes from 22 known Nudibranchia species’ mitochondrial genomes () using MAFFT version 7.508 (Katoh and Standley Citation2013). This was done based on their amino acid sequences, as illustrated in Supplementary Data 1. For each alignment, seven species belonging to the suborder Cladobranchia were selected. The selected 13 alignments were then concatenated. Based on the concatenated alignment, the molecular phylogenetic tree was generated with the maximum likelihood method with 1000 bootstrap replicates in MEGA version 10.1.8 (Kumar et al. Citation2018).
Table 1. The list of species, GenBank accession ID, and references used for alignments (Supplementary Data 1).
Results and discussion
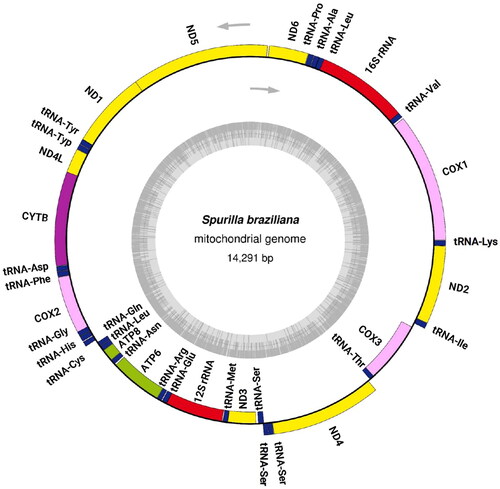
The mitochondrial genome of S. braziliana (GenBank Accession ID: LC759638) is 14,291 bp and has a GC content of 33.94%. It consists of 13 protein-coding genes, two rRNA genes, and 23 tRNA genes (). All 13 protein-coding genes exhibit high amino acid sequence similarity with those of other nudibranch species (Supplementary Data 1), suggesting the accuracy of the genome annotation. The gene order is identical to that of Sakuraeolis japonica (Karagozlu et al. Citation2016a, Citation2016b) and Hermissenda emurai (Dinh Do, Choi, et al. Citation2019; Dinh Do, Kim, et al. Citation2019), both of which belong to the superfamily Aeolidioidea, the same as S. braziliana. The start codons for S. braziliana were compared with those of 22 reference species (with 21 species for ND5 gene). This comparison revealed six different start codons in 23 nudibranchs: ATA, ATT, ATG, ATC, TTG, and GTG. The start codon usage in S. braziliana was found to be distinct, particularly the use of GTG in ATP6, ATP8, COX1, and COX2 genes, a pattern rarely observed in the comparison species (ATP6: 1/23 species, ATP8: 1/23, COX1: 6/23, and COX2: 1/23). Within the NADH dehydrogenase gene series, TTG serves as the start codon in 5 genes, excluding ND4 and ND5. For the remaining 4 genes, COX3, CYTB, and ND4 adopt ATG as their start codon, while ND5 employs ATA. Berghia stephanieae, which belongs to the same family, Aeolidiidae, as S. braziliana, has been reported to exhibit tRNA-Ser 1 gene duplication (Melo Clavijo et al. Citation2021). In our S. braziliana sample as well, gene duplications of tRNA-Ser were observed, and these tRNAs exhibited a unique secondary structure with a missing D-arm (Supplementary Data 2).
Figure 2. Circular map of the complete mitochondrial genome annotated to show the locations and orientation of its 13 protein-coding genes, 2 rRNA genes, and 23 tRNA genes. The upper arrows indicate the direction of transcription, while the inner gray circle represents the GC content.
Through our blastn analyses of the annotated COX1 and 16S rRNA genes, we found that they only exhibited over 90% similarity with two species, S. braziliana and S. neapolitana, thus confirming that our sample belongs to the genus Spurilla. The COX1 gene showed 100% similarity with one of the S. braziliana references, but did not demonstrate complete similarity with the S. neapolitana reference. Given that all Spurilla nudibranchs found in the Pacific are S. braziliana (Carmona et al. Citation2013), we can confidently assert that our specimen is S. braziliana.
As shown in the molecular phylogenetic tree (), S. braziliana forms a monophyletic group with B. stephanieae and members of the superfamily Aeolidioidea.
Figure 3. Maximum likelihood phylogenetic tree of S. braziliana and six other Cladobranchia species based on the concatenated amino acid alignments of the 13 protein-coding genes in the mitochondrial genome. Bootstrap support values are shown for each node, based on 1000 replicates. The list of species and their GenBank accession IDs are shown in .

The previous molecular phylogenetic study of the superfamily Aeolidioidea using COI, 16S rRNA, and H3 genes showed that the monophyletic group containing the genera Spurilla and Berghia was a sister group to the group containing Sakuraeolis (Carmona et al. Citation2011). The phylogenetic tree generated using all the protein-coding genes of the mitochondrial genome matched these findings. The mitochondrial genome of S. braziliana, the first mitochondrial genome of its genus, will provide important data for future taxonomic studies on Spurilla nudibranchs, which are known for their highly variable morphology.
Ethical approval
The ‘Manual for Animal Experiment of the University of Tokyo’ provides ethical guidelines only for mammals, birds, and reptiles, and does not require ethical review for the invertebrates used in this study. Therefore, any specific permission from the ‘Office for Life Science Research Ethics and Safety’ of the University of Tokyo is not necessary for this research.
Author contributions
H.M., R.Y., and K.Y. formulated the research concept. H.M., K.H., and R.Y. collected the S. braziliana samples. H.M. conducted the experimental manipulation and analysis, excluding library preparation and next-generation sequencing tasks. Supervision was provided by S.K., K.Y., and S.A. All authors contributed to data interpretation and discussion of the results. The manuscript was written by H.M. and A.L. All authors participated in the revision of the manuscript draft and approved the final version. All authors agree to be accountable for all aspects of the work and ensure that inquiries concerning the accuracy or integrity of any part of the work are thoroughly investigated and resolved.
Supplemental Material
Download PNG Image (133.4 KB)Supplemental Material
Download MS Power Point (117.6 KB)Supplemental Material
Download MS Power Point (1.2 MB)Supplemental Material
Download MS Power Point (10.5 MB)Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI at https://www.ncbi.nlm.nih.gov under the accession ID: LC759638. The associated BioProject, SRA, and Bio-sample numbers are PRJDB15401, DRR450785, and SAMD00585630, respectively.
Additional information
Funding
References
- Carmona L, Gosliner TM, Pola M, Cervera JL. 2011. A molecular approach to the phylogenetic status of the aeolid genus Babakina roller, 1973 (Nudibranchia). J Molluscan Stud. 77(4):417–422. doi: 10.1093/mollus/eyr029.
- Carmona L, Lei BR, Pola M, Gosliner TM, Valdés Á, Cervera JL. 2014. Untangling the Spurilla neapolitana (Delle Chiaje, 1841) species complex: a review of the genus Spurilla Bergh, 1864 (Mollusca: Nudibranchia: Aeolidiidae). Zool J Linn Soc. 170(1):132–154. doi: 10.1111/zoj.12098.
- Carmona L, Pola M, Gosliner TM, Cervera JL. 2013. A tale that morphology fails to tell: a molecular phylogeny of Aeolidiidae (Aeolidida, Nudibranchia, Gastropoda). PLOS One. 8(5):e63000. doi: 10.1371/journal.pone.0063000.
- Danecek P, Bonfield JK, Liddle J, Marshall J, Ohan V, Pollard MO, Whitwham A. 2021. Twelve years of SAMtools and BCFtools. GigaScience. 10(2):giab008. doi: 10.1093/gigascience/giab008.
- Darty K, Denise A, Ponty Y. 2009. VARNA: interactive drawing and editing of the RNA secondary structure. Bioinformatics. 25(15):1974–1975. doi: 10.1093/bioinformatics/btp250.
- Dinh Do T, Choi TJ, Jung DW, Kim JI, Karagozlu MZ, Kim CB. 2019. The complete mitochondrial genome of Phyllidiella Pustulosa (Cuvier, 1804) (Nudibranchia, Phyllidiidae). Mitochondrial DNA B. 4(1):771–772. doi: 10.1080/23802359.2019.1565976.
- Dinh Do T, Kim JI, Jung DW, Choi TJ, Karagozlu MZ, Kim CB. 2019. Characterization of the complete mitochondrial genome of Hermissenda Emurai (Baba, 1937) (Nudibranchia, Facelinidae). Mitochondrial DNA B. 4(1):860–861. doi: 10.1080/23802359.2019.1572477.
- Do T, Dinh Y, Choi DW, Jung, CB, Kim . 2020. Caution and curation for complete mitochondrial genome from next-generation sequencing: a case study from dermatobranchus otome (Gastropoda, Nudibranchia). Anim Syst Evol Div. 36(4):336–346.
- Do TD, Jung DW, Kim CB. 2022. Molecular phylogeny of selected dorid nudibranchs based on complete mitochondrial genome. Sci Rep. 12(1):18797. doi: 10.1038/s41598-022-23400-9.
- Donath A, Jühling F, Al-Arab M, Bernhart SH, Reinhardt F, Stadler PF, Middendorf M, Bernt M. 2019. Improved annotation of protein-coding genes boundaries in metazoan mitochondrial genomes. Nucleic Acids Res. 47(20):10543–10552. doi: 10.1093/nar/gkz833.
- Grande C, Templado J, Cervera JL, Zardoya R. 2004. Phylogenetic relationships among opisthobranchia (Mollusca: Gastropoda) based on mitochondrial Cox 1, TrnV, and RrnL genes. Mol Phylogenet Evol. 33(2):378–388. doi: 10.1016/j.ympev.2004.06.008.
- Greenwood PG, Mariscal RN. 1984. The utilization of cnidarian nematocysts by aeolid nudibranchs: nematocyst maintenance and release in spurilla. Tissue Cell. 16(5):719–730. doi: 10.1016/0040-8166(84)90005-3.
- Greiner S, Lehwark P, Bock R. 2019. OrganellarGenomeDRAW (OGDRAW) version 1.3.1: expanded toolkit for the graphical visualization of organellar genomes. Nucleic Acids Res. 47(W1):W59–W64. doi: 10.1093/nar/gkz238.
- Gruber AR, Lorenz R, Bernhart SH, Neubock R, Hofacker IL. 2008. The Vienna RNA websuite. Nucleic Acids Res. 36:W70–W74. doi: 10.1093/nar/gkn188.
- Karagozlu MZ, Sung JM, Lee J, Kim SG, Kim CB. 2016a. Complete mitochondrial genome analysis of Sakuraeolis Japonica (Baba, 1937) (Mollusca, Gastropoda, Nudibranchia). Mitochondrial DNA B Resour. 1(1):720–721. doi: 10.1080/23802359.2016.1229587.
- Karagozlu MZ, Sung J, Lee J, Kwak W, Kim CB. 2016b. Complete sequences of mitochondrial genome of Hypselodoris Festiva (A. Adams, 1861) (Mollusca, Gastropoda, Nudibranchia). Mitochondrial DNA B Resour. 1(1):266–267. doi: 10.1080/23802359.2016.1159933.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780. doi: 10.1093/molbev/mst010.
- Kim H, Yoon M, Kim KY, Jung YH. 2021. The complete mitochondrial genome of sea slug Phyllidiopsis Krempfi Pruvot-Fol, 1957 (Nudibranchia, Phyllidiidae) from Pacific Ocean. Mitochondrial DNA B. 6(4):1523–1524. doi: 10.1080/23802359.2020.1823898.
- Kumar S, Stecher G, Li M, Knyaz C, Tamura K. 2018. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. 35(6):1547–1549. doi: 10.1093/molbev/msy096.
- Li H. 2013. Aligning sequence reads. Clone sequences and assembly contigs with BWA-MEM. arXiv:1303.3997v2 doi: 10.48550/ARXIV.1303.3997.
- Medina M, Lal S, Vallès Y, Takaoka TL, Dayrat BA, Boore JL, Gosliner T. 2011. Crawling through time: transition of snails to slugs dating back to the Paleozoic, based on mitochondrial phylogenomics. Mar Genomics. 4(1):51–59. doi: 10.1016/j.margen.2010.12.006.
- Melo Clavijo J, Drews F, Pirritano M, Simon M, Salhab A, Donath A, Frankenbach S, Serôdio J, Bleidißel S, Preisfeld A, et al. 2021. The complete mitochondrial genome of the photosymbiotic sea slug Berghia Stephanieae (Valdés, 2005) (Gastropoda, Nudibranchia). Mitochondrial DNA B Resour. 6(8):2281–2284. doi: 10.1080/23802359.2021.1914211.
- Meng G, Li Y, Yang C, Liu S. 2019. MitoZ: a toolkit for animal mitochondrial genome assembly, annotation and visualization. Nucleic Acids Res. 47(11):e63–e63. doi: 10.1093/nar/gkz173.
- Sevigny JL, Kirouac LE, Thomas WK, Ramsdell JS, Lawlor KE, Sharifi O, Grewal S, Baysdorfer C, Curr K, Naimie AA, et al. 2015. The mitochondrial genomes of the Nudibranch Mollusks, Melibe Leonina and Tritonia Diomedea, and their impact on gastropod phylogeny. PLOS One. 10(5):e0127519. doi: 10.1371/journal.pone.0127519.
- Walker BJ, Abeel T, Shea T, Priest M, Abouelliel A, Sakthikumar S, Cuomo CA, Zeng Q, Wortman J, Young SK, et al. 2014. Pilon: an integrated tool for comprehensive microbial variant detection and genome assembly improvement. PLOS One. 9(11):e112963. doi: 10.1371/journal.pone.0112963.
- Yu C, Kim H, Kim HJ, Jung YH. 2018. The complete mitochondrial genome of the oriental sea slug: Chromodoris orientalis (Nudibranchia, Chromodorididae). Mitochondrial DNA B Resour. 3(2):1017–1018. doi: 10.1080/23802359.2018.1508381.
