Abstract
Helicana japonica mainly inhabits burrowed holes in the mudflats and intertidal zones. Specimens from the Republic of Korea were collected and whole genomic DNA from the cheliped muscle tissue was extracted. We determined the complete mitochondrial genome using Illumina HiSeq X Ten. The mitogenome is 16,535 bp in length and consists of 13 protein-coding genes, 2 rRNA genes, and 22 tRNA genes. A phylogenetic tree was reconstructed using the maximum-likelihood of phylogeny methods. H. japonica formed a sister clade with Helicana wuana, which is another Helicana species.
Introduction
Helicana japonica (Sakai and Yatsuzuka Citation1980) (Crustacea: Decapoda: Varunidae) inhabits the muddy banks of estuaries and salt marshes from southern Japan to Korea and Taiwan (Irawan and Kijima, Citation1994; Omori et al. Citation2006; Shih and Suzuki Citation2008) and Helicana wuana inhabits a similar environment. However, the morphological differences between the two Helicana species were unclear. Therefore, it is necessary to study the genetic characteristics that distinguish the two species. This study reported the first complete mitogenome of H. japonica and will provide genetic information for improving the taxonomy of Helicana.
Materials and methods
H. japonica specimens were collected from Daesan-eup, Seosan-si, Chungcheongnam-do, Republic of Korea (36°58'6.67"N, 126°20'14.80"E). Total genomic DNA was extracted from the specimens using a DNeasy Blood & Tissue DNA Kit (Qiagen, Hilden, Germany). A genomic library was constructed using the QIAseq FX Single Cell DNA Library kit (Qiagen, Hilden, Germany) with paired-end reads. Next-generation sequencing analysis was performed using Illumina HiSeq X Ten (Illumina Inc., San Diego, CA, USA). The quality of all sequence was checked using FastQC v. 0.11.5 (Andrews, Citation2010) to obtain clean data, and reads were trimmed with Trimmomatic v. 0.36 (Bolger et al. Citation2014). De novo assembly was performed using SPAdes 3.13.0. (Bankevich et al. Citation2012). Mitochondrial genes were predicted and annotated using MitoZ v. 2.3 (Meng et al. Citation2019). The definitive species information was genetically identified by cytochrome oxidase subunit I (COI) from the mitochondrial genome results. For species identification, COI sequences of the related species were aligned using Geneious Prime v. 2022.1.1 (Biomatters, Auckland, New Zealand), and calculated genetic variations using Mega X (Kumar et al. Citation2018) with the Kimura two-parameter model (Kimura, Citation1980). To investigate the phylogenetic status of Helicana japonica in Varunidae, combined 10,795 bp sequences of the 12 protein-coding genes (PCGs) from 11 Brachyura species and an outgroup species were compared. The species sequence data were downloaded from NCBI GenBank, and PCG ND6 was excluded because of its loss in some species. The PCGs sequences were aligned by MAFFT v.7.490 (Katoh and Standley, Citation2013), a maximum-likelihood phylogenetic tree was reconstructed using Phy ML 3.0 (Guindon et al. Citation2010) with 1000 bootstraps. The best model selected using PAUP* v.4a168 (Swofford, Citation2003) was the GTR + G + I model. The specimens were preserved in ethanol and deposited in the National Marine Biodiversity Institute of Korea (https://www.mabik.re.kr, Ji Min Kim, and [email protected], Voucher No. MABIK CR00248070).
Results and discussion
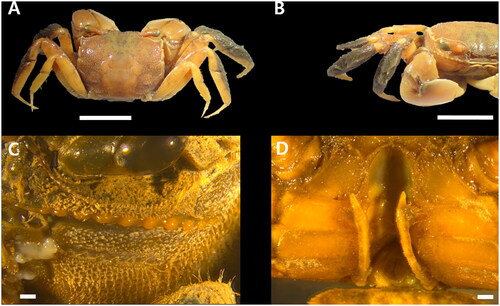
The morphological identification of H. japonica was based on descriptions by Kim (Citation1973) and Sakai et al. (Citation2006) (). As described in the references, the suborbital ridge of the male consisted of a row of 10 tubercles, and the last tooth hardly developed (). The first and second ambulatory legs comprised a thick pelt of setae, surfaces of the third ambulatory leg with setae, and the fourth ambulatory leg without setal tufts (). The first pleopods were slender, stem incurved distally, and swollen in the middle ().
Figure 1. Helicana japonica male–Scale 10 mm (A, B); 1 mm (C, D). (A) dorsal over view; (B) Ambulatory legs; (C) Suborbital ridge; (D) First pleopods. (photos taken by Ji Min Kim at MABIK).
In the 658 bp COI comparison for molecular identification, the H. japonica presented in this study was 100% identical with the Japanese Helicana japonica (AB334553) of (Shih & Suzuki (Citation2008) and 99.8% identical with the Korean H. japonica (JX502897) (intraspecific variation 0.000 ∼ 0.002). Furthermore, comparison with allied species in related genera confirmed a genetic differences (interspecific variation 0.058 ∼ 0.215; ). During the blasting process for the molecular comparison, we found that a Helicana wuana sequence already registered in NCBI (NC034995/KX334898; Tang et al. Citation2018) matched ours. However, this data was consequently excluded from the present comparative analysis due to taxonomic uncertainty because: (1) it showed genetic differences from other H.wuana sequences (MH593562, JX502899, AB334551); and (2) no morphological description of the specimens was provided.
Table 1. Pairwise genetic distances based on 658 bp size of COI genes from H. japonica. (*Outgroup).
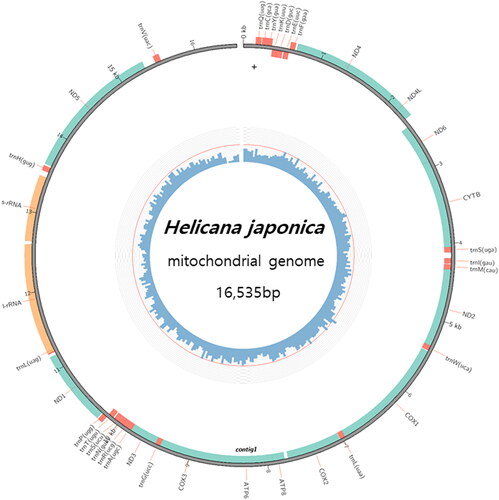
The complete mitochondrial gene sequence of H. japonica is a circular molecule of 16,535 bp consisting of 13 protein-coding genes (PCGs), 2 ribosomal RNA (rRNA) genes, and 22 tRNA genes (). The full-length mitochondrial genome sequence was deposited in GenBank accession number ON646695. The overall nucleotide composition was A (33.0%), T (35.6%), C (19.8%), G (11.6%) and GC content (31.4%). Most genes were encoded by the heavy strand, and only ND4, ND4L, ND1, ND5, 16S rRNA, 12S rRNA tRNA-Gln, tRN-Cys, tRNA-Tyr, tRNA-Phe, tRNA-Pro, tRNA-Leu, tRNA-His, and tRNA-Val were encoded by the light strand. There are four types of start codons: ATG (ND4, ND4L, CYTB, COX1, COX2, ATP8, COX3, ND5), ATT (ND6, ATP6), ATC (ND2, ND3), ATA (ND1). There are also two types of stop codons: TAA (ND4L, ND6, COX1, ATP8, ATP6, COX3, ND3, ND5), TAG (ND4, CYTB, ND2, COX2, ND1).
Figure 2. Complete mitochondrial genome circle map of Helicana japonica. The genome is 16,535 bp consisting of 13 PCGs, 2 rRNA, and 22 tRNA genes.
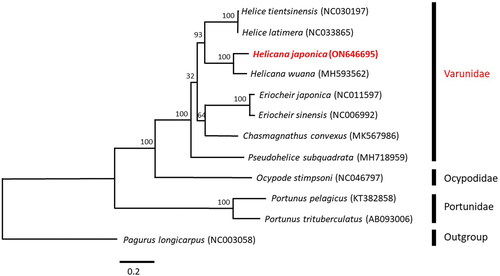
The phylogenetic analysis result showed that the family Varunidae is a monophyly group. Within the clade, Varunidae species were subdivided into five clades: Helice, Helicana, Eriocheir, Chasmagnathus, and Pseudohelice. Similar to the findings of Shih and Suzuki (Citation2008), the phylogenetic tree revealed that H. japonica formed a sister clade with H. wuana, another Helicana species ().
Figure 3. Phylogenetic tree of maximum-likelihood (ML) phylogenetic trees based on the mitochondrial 12 protein-coding genes (PCGs). the values of bootstrap support (ML) is shown the nodes. The following sequences were used: Helice tientsinensis NC030197 (identical to KR336555; Xin et al. Citation2017), Helice latimera NC033865 identical to KU589291; Tang et al. Citation2020), Helicana wuana MH593562 (Yuan et al. Citation2018), Eriocheir japonica NC011597 (identical to FJ455505; Wang et al. Citation2016), Eriocheir sinensis NC006992 (Sun et al. Citation2005), Chasmagnathus convexus MK567986 (Guo et al. Citation2019), Pseudohelice subquadrata MH718959 (Kim et al. Citation2019), ocypode stimpsoni NC046797 (identical to MN917464; Kim and Jung, Citation2020), Portunus pelagicus KT382858 (Koolkarnkhai et al. Citation2019), Portunus trituberculatus AB093006 (Yamauchi et al. Citation2003), and pagurus longicarpus NC003058 (identical to AF150756; Hickerson and Cunningham, Citation2000); (11 species sequence data were downloaded from NCBI).
Authors’ contributions
This study was designed and conceived of by JMK and CHY. JMK and HSK collected and identified the materials. JMK and CHY contributed significantly to phylogenetic analysis and manuscript preparation. HSK was involved in the interpretation of data and critically revised the manuscript for intellectual content. All authors approved the final version to be published and agreed to be accountable for all aspects of this work.
Ethical approval
The samples collected and used in this study did not include any marine organisms under protection, as determined by the Ordinance of the Ministry of Oceans and Fisheries in the Republic of Korea. Therefore, our study was exempted from ethical approval and did not require any permission to conduct it.
Geolocation information
Daesan-eup, Seosan-si, Chungcheongnam-do, Republic of Korea (36°58'6.67" N, 126°20'14.80" E).
Supplemental Material
Download PNG Image (130.1 KB)Disclosure statement
No potential conflict of interest was reported by the authors.
Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank(https://www.ncbi.nlm.nih.gov/) under the accession no. ON646695. The associated BioProject, SRA, and Bio-Sample numbers were PRJNA785076, SRR17084632, and SAMN23526795, respectively.
Additional information
Funding
References
- Andrews S. 2010. FastQC: a quality control tool for high throughput sequence data. (http://www.bioinformatics.babraham.ac.uk/projects/fastqc/).
- Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, et al. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 19(5):455–477. doi: 10.1089/cmb.2012.0021.
- Bolger AM, Lohse M, Usadel B. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 30(15):2114–2120. http://www.usadellab.org/cms/?page=trimmomatic. doi: 10.1093/bioinformatics/btu170.
- Guindon S, Dufayard J-F, Lefort V, Anisimova M, Hordijk W, Gascuel O. 2010. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol. 59(3):307–321. doi: 10.1093/sysbio/syq010.
- Guo Y, Feng J, Ye Y, Li J, Guo B, Xie J, Zhu A. 2019. The complete mitochondrial genome and phylogenetic analysis of Chasmagnathus convexus (Crustacea: Grapsidae). Mitochondrial DNA Part B. 4(1):1652–1653. doi: 10.1080/23802359.2019.1607582.
- Hickerson MJ, Cunningham CW. 2000. Dramatic mitochondrial gene rearrangements in the hermit crab Pagurus longicarpus (Crustacea, Anomura). Mol Biol Evol. 17(4):639–644. doi: 10.1093/oxfordjournals.molbev.a026342.
- Irawan B, Kijima A. 1994. Difference of salinity requirements among the three estuarine crab species, Chiromantes dehaani, Helice tridens and H. japonica (Brachyura: Grapsidae). Tohoku J Agri Res. 44:1–4.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780. doi: 10.1093/molbev/mst010.
- Kim HS. 1973. Illustrated encyclopedia of fauna and flora of Korea. Vol. 14. Anomura: Brachyura. Seoul: Samwha Publishing co. p. 1–649.
- Kim HS, Kim KY, Lee SH, Hong SS, Cho IY, Yi CH, Kim IH, Yoon M, Kim MS. 2019. The complete mitochondrial genome of Pseudohelice subquadrata (Dana, 1851) (Crustacea: decapoda: varunidae). Mitochondrial DNA Part B. 4(1):103–104. doi: 10.1080/23802359.2018.1536491.
- Kim H, Jung J. 2020. Complete mitochondrial genome of the ghost crab Ocypode stimpsoni Ortmann, 1897 (Brachyura: Decapoda: Ocypodidae) and its phylogenetic relationship in Brachyura. Mitochondrial DNA Part B. 5(2):1699–1700. doi: 10.1080/23802359.2020.1749160.
- Kimura M. 1980. A simple method of estimating evolutionary rate of base substitution through comparative studies of nucleotide sequences. J Mol Evol. 16(2):111–120. doi: 10.1007/BF01731581.
- Koolkarnkhai P, Intakham C, Sangthong P, Surat W, Wonnapinij P. 2019. Portunus pelagicus mtDNA heteroplasmy inheritance and its effect on the use of mtCR and mtCOI sequence data. Mitochondrial DNA A DNA Mapp Seq Anal. 30(8):848–860. doi: 10.1080/24701394.2019.1693549.
- Kumar S, Stecher G, Li M, Knyaz C, Tamura K. 2018. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. 35(6):1547–1549. doi: 10.1093/molbev/msy096.
- Meng G, Li Y, Yang C, Liu S. 2019. MitoZ: a toolkit for mitochondrial genome assembly, annotation and visualization. Nucleic Acids Res. 47(11):e63. doi: 10.1093/nar/gkz173.
- Omori K, Kikutani Y, Irawan B, Goda Y. 2006. Size-dependent intraguild reciprocal predation between Helice tridens De Haan and H. japonica Sakai and Yatsuzuka (Decapoda: Grapsidae) as analyzed in field experiments. J Crustacean Biol. 26(2):148–153. doi: 10.1651/S-2645.1.
- Sakai K, Turkay M, Yang SL. 2006. Revision of the Helice/Chasmagnathus complex (Crustacea: dedapoda: brachyura). Abh Senckb Naturforsch Ges. 565:1–76.
- Sakai K, Yatsuzuka K. 1980. Notes on some Japanese and Chinese Helice with Helice (Helicana) n. subgen., including Helice (Helicana) japonica n. sp. (Crustacea: Decapoda). Senckenbergia Biologica. 60(5–6): 393–411.
- Shih HT, Suzuki H. 2008. Taxonomy, phylogeny, and biogeography of the endemic mudflat crab Helice/Chasmagnathus complex (Crustacea: brachyura: varunidae) from East Asia. Zool Stud. 47(1):114–125.
- Sun H, Zhou K, Song D. 2005. Mitochondrial genome of the Chinese mitten crab Eriocheir japonica sinenesis (Brachyura: Thoracotremata: Grapsoidae) reveals a novel gene order and two target regions of gene rearrangements. Gene. 349:207–217. doi: 10.1016/j.gene.2004.12.036.
- Swofford DL. 2003. PAUP*: phylogenetic analysis using parsimony (*and other methods), vers. 4. Sunderland, MA: Sinauer Associates.
- Tang BP, Liu Y, Xin ZZ, Zhang DZ, Wang ZF, Zhu XY, Wang Y, Zhang HB, Zhou CL, Chai XY, et al. 2018. Characterisation of the complete mitochondrial genome of Helice wuana (Grapsoidea: varunidae) and comparison with other Brachyuran crabs. Genomics. 110:221–230. doi: 10.1016/j.ygeno.2017.10.001.
- Tang YY, Tang BP, Xin ZZ, Li YT, Zha XH, Zhang DZ, Sun Y, Liu QN, Ma YF. 2020. Characterization of the complete mitochondrial genome of Helice latimera and its phylogenetic implications in Brachyura. Genomics. 112(6):5180–5187. doi: 10.1016/j.ygeno.2020.08.013.
- Wang J, Huang L, Cheng Q, Lu G, Wang C. 2016. Complete mitochondrial genomes of three mitten crabs, Eriocheir sinensis, E. hepuensis, and E. japonica. Mitochondrial DNA A DNA Mapp Seq Anal. 27(2):1175–1176. doi: 10.3109/19401736.2014.936425.
- Xin ZZ, Liu Y, Zhang DZ, Wang ZF, Zhang HB, Tang BP, Zhou CL, Chai XY, Liu QN. 2017. Mitochondrial genome of Helice tientsinensis (Brachyura: Grapsoidea: Varunidae): gene rearrangements and higher-level phylogeny of the Brachyura. Gene. 627(5): 307–314. doi: 10.1016/j.gene.2017.06.036.
- Yamauchi MM, Miya MU, Nishida M. 2003. Complete mitochondrial DNA sequence of the swimming crab, Portunus trituberculatus (Crustacea: Decapoda: Brachyura). Gene. 311:129–135. doi: 10.1016/s0378-1119(03)00582-1.
- Yuan Y, He Y, Liu S, Ji X, Qin Y, Wang X. 2018. The complete mitochondrial genome of Helice sheni and its phylogenetic implication. Mitochondrial DNA B Resour. 3(2):1183–1184. doi: 10.1080/23802359.2018.1524721.
