Abstract
In this study, the complete mitochondrial genome of the Taiwan tai Argyrops bleekeri was determined for the first time by next-generation sequencing. The circular mtDNA molecule was 16,646 bp in size and the overall base composition was A (27.77%), C (28.95%), G (16.60%), and T (26.68%), with a slight bias toward A + T. The complete mitogenome encoded 13 protein-coding genes (PCGs), 22 tRNA genes, two rRNA genes, and a control region. Phylogenetic analysis based on the 13 PCGs of the Sparidae family revealed that Argyrops appears to be most closely related to Pagrus and Parargyrops, but further research is needed.
Introduction
Argyrops bleekeri Oshima, 1927, commonly known as Taiwan tai, is widely distributed in the western Pacific from southern Japan to northwestern Australia (Uehara and Tachihara Citation2020). It usually inhabits sandy and muddy bottoms, and is an important target for regional fisheries. The genus of Argyrops, with multiple elongated and filamentous dorsal fin spines, was long thought to have only four valid species worldwide until Iwatsuki and Heemstra (Citation2018) reported three new species from the Indo-West Pacific. Argyrops bleekeri is distinguished from its congeners in having only a rudimentary first dorsal fin spine and usually five elongated dorsal fin spines () (Iwatsuki and Heemstra Citation2018). So far, there is still a gap in the complete sequence of the mitochondrial genome of the Argyrops genus (Caputi et al. Citation2021; Pan et al. Citation2021; Kawai et al. Citation2022; Osca et al. Citation2022). The study of the phylogeny of the genus Argyrops is severely hampered by a lack of data. In this study, we sequenced and assembled the mitochondrial genome of A. bleekeri, the first for the genus Argyrops. The result will be useful for species identification and phylogenetic studies within Sparidae.
Materials and methods
One specimen was captured from East China Sea (27.516°N, 124.517°E) in August 2020. It was deposited in the East China Sea Fisheries Research Institute, Chinese Academy of Fishery Sciences with the voucher number 2106010064 (Hanye Zhang, [email protected]). The genomic DNA was extracted from muscle tissue, which was sequenced by Illumina NovaSeq 6000 platform with both directions of 150 bp reads at Personal Biotechnology Co., Ltd. (Shanghai, China). The genome was assembled de novo with A5-miseq v20150522 (Coil et al. Citation2015) and SPAdes v3.9.0 (Bankevich et al. Citation2012). The read coverage depth map is shown in Figure S1. The complete mitogenome was annotated using online tool MitoAnnotator (Iwasaki et al. Citation2013). The annotated sequence was submitted to GenBank with accession number MZ892909.
The circular genome map was generated using the OGDRAW version 1.3.1 (Greiner et al. Citation2019). A phylogenetic tree was constructed to find the phylogenetic position of A. bleekeri. Twenty-six other published, complete mitogenome sequences of the Sparidae family were obtained from GenBank. Thirteen protein-coding genes (PCGs) were extracted from the annotated sequences and aligned using MAFFT version 7 (Katoh and Standley Citation2013). The resulting alignment was then subjected to maximum-likelihood (ML) analyses performed using IQ-TREE v1.6.8 (Nguyen et al. Citation2015) for 5000 ultrafast (Minh et al. Citation2013) bootstraps. The best partitioning scheme and nucleotide substitution model, GTR + I + G, were determined with PartitionFinder2 (Lanfear et al. Citation2017).
Results and discussion
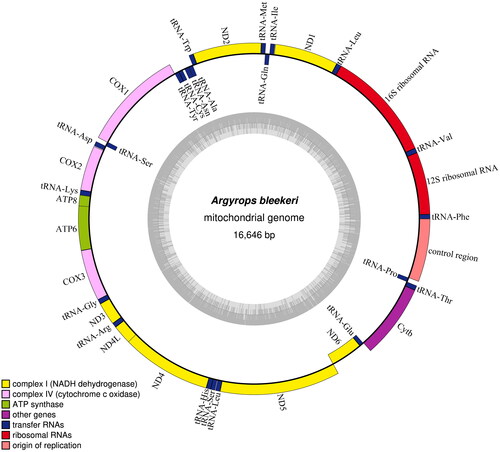
The complete circular mitochondrial genome of A. bleekeri is 16,646 bp in length (), showing a base composition of: A 27.77%, C 28.95%, G 16.60%, and T 26.68%, with a slight bias toward A + T, which is similar to other Sparidae species (Celestina et al. Citation2019). Its mitogenome contains two rRNA genes, 13 PCGs, 22 tRNA genes, and a control region. All PCGs begin with an ATG start codon except for COI with GTG. COI terminates with AGG as stop codon; ND1 with TAG; ATP8, ND4L, ND5, and ND6 with TAA; other seven PCGs end with an incomplete stop codon TA– or T––. Except for ND6 gene and eight tRNA genes (tRNA-Gln, tRNA-Ala, tRNA-Asn, tRNA-Cys, tRNA-Tyr, tRNA-Ser, tRNA-Glu, and tRNA-Pro), all other genes were located on the heavy strand (H-strand).
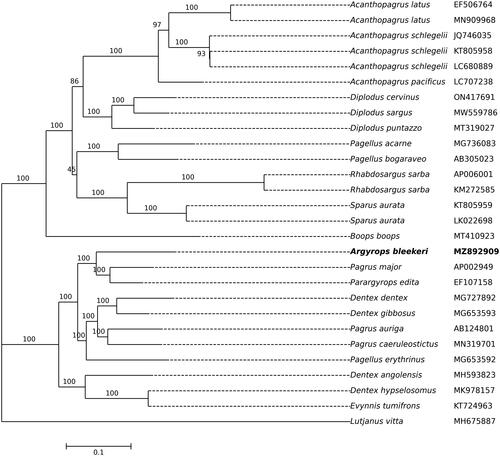
In the absence of other Argyrops species, the inferred phylogenetic tree suggests that Argyrops appears to be most closely related to Pagrus and Parargyrops, with strong bootstrap support (). However, the three species of the Pagrus and the four species of the Dentex do not individually cluster into monophyletic groups. It also suggested that the relationships in the Sparidae may need further investigation.
Ethical approval
The fish were collected during the fisheries resource survey for which permission was granted by Ministry of Agriculture and Rural Affairs of the People’s Republic of China.
Author contributions
H. Zhang and J. Ling designed the study and wrote the manuscript. X. Song and Z. Liu carried out the experiments and analyzed the data. J. Ling performed all fieldwork and sampling. H. Zhang and X. Song revised the manuscript. All authors revised the manuscript and agree to be responsible for all aspects of the work.
Supplemental Material
Download MS Word (74.3 KB)Disclosure statement
No potential conflict of interest was reported by the authors. The authors alone are responsible for the content and writing of this article.
Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI at https://www.ncbi.nlm.nih.gov/ under the accession number MZ892909. The associated BioProject, SRA, and BioSample numbers are PRJNA756399, SRR15534453, and SAMN20865296, respectively.
Additional information
Funding
References
- Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, et al. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 19(5):455–477. doi: 10.1089/cmb.2012.0021.
- Caputi L, Osca D, Ceruso M, Venuti I, Sepe RM, Anastasio A, D'Aniello S, Crocetta F, Pepe T, Sordino P. 2021. The complete mitochondrial genome of the white seabream Diplodus sargus (Perciformes: Sparidae) from the Tyrrhenian Sea. Mitochondrial DNA B Resour. 6(9):2581–2583. doi: 10.1080/23802359.2021.1915209.
- Celestina M, Marina C, Claudia C, Giuseppe P, Aniello A, Paolo S, Tiziana P. 2019. The complete mitochondrial genome of the Angolan dentex Dentex angolensis (Perciformes: Sparidae). Mitochondr DNA B. 4(1):1245–1246.
- Coil D, Jospin G, Darling A. 2015. A5-miseq: an updated pipeline to assemble microbial genomes from Illumina MiSeq data. Bioinformatics. 31(4):587–589. doi: 10.1093/bioinformatics/btu661.
- Greiner S, Lehwark P, Bock R. 2019. OrganellarGenomeDRAW (OGDRAW) version 1.3.1: expanded toolkit for the graphical visualization of organellar genomes. Nucleic Acids Res. 47(W1):W59–W64. doi: 10.1093/nar/gkz238.
- Iwasaki W, Fukunaga T, Isagozawa R, Yamada K, Maeda Y, Satoh TP, Sado T, Mabuchi K, Takeshima H, Miya M, et al. 2013. MitoFish and MitoAnnotator: a mitochondrial genome database of fish with an accurate and automatic annotation pipeline. Mol Biol Evol. 30(11):2531–2540. doi: 10.1093/molbev/mst141.
- Iwatsuki Y, Heemstra PC. 2018. Taxonomic review of the genus Argyrops (Perciformes; Sparidae) with three new species from the Indo-West Pacific. Zootaxa. 4438(3):401–442. doi: 10.11646/zootaxa.4438.3.1.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780. doi: 10.1093/molbev/mst010.
- Kawai K, Fujita H, Umino T. 2022. The complete mitochondrial genome of Acanthopagrus pacificus (Perciformes, Sparidae) from Iriomotejima Island, Okinawa, Japan. Mitochondrial DNA B Resour. 7(6):1027–1029. doi: 10.1080/23802359.2022.2080606.
- Lanfear R, Frandsen PB, Wright AM, Senfeld T, Calcott B. 2017. PartitionFinder 2: new methods for selecting partitioned models of evolution for molecular and morphological phylogenetic analyses. Mol Biol Evol. 34(3):772–773. doi: 10.1093/molbev/msw260.
- Minh BQ, Nguyen MAT, von Haeseler A. 2013. Ultrafast approximation for phylogenetic bootstrap. Mol Biol Evol. 30(5):1188–1195. doi: 10.1093/molbev/mst024.
- Nguyen LT, Schmidt HA, von Haeseler A, Minh BQ. 2015. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 32(1):268–274. doi: 10.1093/molbev/msu300.
- Osca D, Caputi L, Tanduo V, Sepe RM, Liberti A, Tiralongo F, Venuti I, Ceruso M, Crocetta F, Sordino P, et al. 2022. The complete mitochondrial genome of the zebra seabream Diplodus cervinus (Perciformes, Sparidae) from the Mediterranean Sea. Mitochondrial DNA B Resour. 7(11):2006–2008. doi: 10.1080/23802359.2022.2145174.
- Pan C, Gao C, Chen T, Chen X, Yang C, Zeng D, Feng P, Jiang W, Peng M. 2021. The complete mitochondrial genome of yellowfin seabream, Acanthopagrus latus (Percoiformes, Sparidae) from Beibu Bay. Mitochondrial DNA B Resour. 6(4):1313–1314. doi: 10.1080/23802359.2021.1907804.
- Uehara M, Tachihara K. 2020. Early development and occurrence patterns of the Pagrinae seabream Argyrops bleekeri (Perciformes: Sparidae): does early life history have implications for sparid relationships? J Fish Biol. 96(3):631–641. doi: 10.1111/jfb.14249.