Abstract
Vaccinium species have great significance as fruit crops due to their economic and food values. Here we report the chloroplast genome of V. oxycoccos. The chloroplast genome of V. oxycoccos was 177,088 bp in length with a GC content of 36.74%. LSC, SSC, and IR regions were 104,139 bp, 3031 bp, and 34,959 bp in length, respectively. The chloroplast genome contained 105 different genes, including 73 protein-coding genes, 4 rRNA genes, and 28 tRNA genes. The phylogenetic analysis indicated that V. oxycoccos was closely related to V. microcarpum in the family Ericaceae. This chloroplast genome not only enriches the genome information of Vaccinium, but also will be useful in the evolution study of the family Ericaceae.
Keywords:
Background
The genus Vaccinium L. (Ericaceae) has about 450 species, occurring mostly in the Northern Hemisphere and in mountainous regions of tropical Asia, Central and South America (Fahrenkrog et al. Citation2022). The genus Vaccinium is diverse and complex, and it has been classified into over 33 subgenera or lineages (Sleumer Citation1941). The Vaccinium taxonomy has been extensively discussed in many literatures, showing the challenging definition of its classification due to morphological overlap between species, different chromosome ploidy, and frequent interspecific hybridization events (Hancock et al. Citation2008). Vaccinium oxycoccos Linnaeus (1753), also called European cranberry, is native to temperate and boreal regions of Europe and North America, also found in Jilin Province, China. V. oxycoccos fruits are rich in significant amounts of vitamin C, anthocyanins, and flavonoids, which have a variety of health effects, including anti-oxidant, anti-inflammatory, hypoglycemic, and hypolipidemic (Ermis et al. Citation2015; Jurikova et al. Citation2018).
Some studies had performed phylogenetic analyses of the Vaccinium using a small amount of molecular data, such as matK, ndhF, nrITS (Kron et al. Citation2002; Powell and Kathleen Citation2002; Schlautman et al. Citation2017). While these studies supported the existence of multiple lineages in Vaccinium, they did not establish and clarify the intimate relationships among the species themselves. Phylogenetic studies of chloroplast genomes are now becoming more common, and chloroplast-based molecular markers are particularly useful for rapid identification of berry products, enabling rapid identification of adulterated berries (Daniell et al. Citation2021). To date, only a few chloroplast genomes of Vaccinium species have been published, while the chloroplast genome of V. oxycoccos remains unknown.
Consequently, we intend to provide a valuable resource for future taxonomic and domestication studies of Vaccinium by assembling and analyzing chloroplast genome of V. oxycoccos. To achieve this goal, we assembled the chloroplast genome of V. oxycoccos and performed comparative genomic analyses with other Vaccinium species. Our primary objectives were to study the feature of the chloroplast genome of V. oxycoccos and to determine its phylogenetic relationship.
Materials and methods
Plant material, DNA extraction and sequencing
Dry leaves of V. oxycoccos were collected in this study from Changbai Mountain, Jilin, China (, 128.094687°E, 42.053095°N). Our experimental studies, including the collection of plant material, were in accordance with institutional, national or international guidelines. The sample was deposited at the herbarium of the College of Pharmaceutical Engineering, Xinyang Agriculture and Forestry University (voucher number: VO001, Xinrong Qiao, [email protected]). Total genomic DNA was extracted using the CTAB method (Doyle and Doyle Citation1987). The DNA library of next generation sequencing with an insert size of 300 bp was constructed and sequenced using the Illumina HiSeq 2000 platform, yielding ∼5 Gb of raw data, and low-quality sequences were removed to obtain clean data.
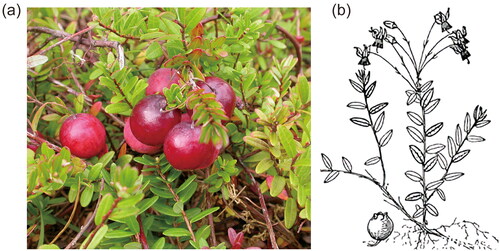
Figure 1. Species reference map of V. oxycoccos. (a) Fruits and leaves of V. oxycoccos (voucher number: VO001; this picture was taken by Xinrong Qiao from the Xinyang Agriculture and Forestry University, Xinyang city, Henan Province, China; 128.094687°E, 42.053095°N;). (b) Sketch of V. oxycoccos (from the Plant photo bank of China). core features: V. oxycoccos is an evergreen subshrub of the genus Vaccinium in the family Ericaceae. Its young branches are slightly hairy, and the stem bark is striped; the leaves are oblong or ovate, dark green above, grayish-white below, and glabrous on both surfaces; the flowers are subumbellate; the pedicels are fine, pilose, and recurved at the tip; the corolla is light red, divided nearly to the base, and the lobes are oblong; the berries are red; the flowering period is June to July; the fruiting period is July to August, and the fruit is edible. Its chromosome number is 2n = 48, and it is a widespread species in the Sub-boreal and boreal zones of the Northern Hemisphere, born in cold swamps, and generally reproduced by seed. In China, it is mainly found in the Changbai Mountain of Jilin Province.
Genome assembly and annotation
De novo genome assembly from the clean data was performed using GetOrganelle v.1.7.5 (Jin et al. Citation2020). The parameters applied for the plastome were “-R 25 -k 21,45,65,85,105,127 -F embplant_pt.” Samtools v1.7 (Li et al. Citation2009) and bedtools v2.28 (Quinlan and Hall Citation2010) were used for depth detection. The chloroplast genome was annotated using CPGAVAS2 (Shi et al. Citation2019), PGA (Qu et al. Citation2019) and Geneious Prime v. 2022.2.2 with a reference genome (V. oldhamii, GenBank: MK049537). GB2sequin (https://chlorobox.mpimp-golm.mpg.de/GenBank2Sequin.html) was then used to confirm the annotation results (Tillich et al. Citation2017). CPGView (Liu et al. Citation2023) was used to test the accuracy of cis- and trans-splicing genes. Visualization of chloroplast genome with OGDraw (Greiner et al. Citation2019) (https://chlorobox.mpimp-golm.mpg.de/OGDraw.html). Comparative sequence hotspot analysis of the chloroplast genomes of Vaccinium species using mVISTA (Frazer et al. Citation2004) (https://genome.lbl.gov/vista/mvista/submit.shtml).
Repeat and IR boundary analysis
Simple sequence repeats (SSRs) were identified using the MISA software (Beier et al. Citation2017), including mono-, di-, tri-, tetra-, penta-, and hexa-nucleotides with minimum numbers of 10, 5, 4, 3, 3, and 3, respectively (Beier et al. Citation2017). Additionally, REPuter (https://bibiserv.cebitec.uni-bielefeld.de/reputer/) was used to calculate palindromic, forward, reverse, and complementary repeats with the following settings: minimum repeat size of 30 bp (Kurtz et al. Citation2001). Furthermore, comparisons between the boundaries of three regions (IR, SSC, LSC) were generated using IRscope (Amiryousefi et al. Citation2018).
Phylogenetic analysis
We phylogenetically analyzed the chloroplast genome of V. oxycoccos with 15 other Ericaceae species. We extracted 74 common protein-coding genes (PCGs) from the genome annotation files using PhyloSuite v. 1.2.2 (Zhang et al. Citation2020). Each gene was aligned using MAFFT v. 7.4 (Katoh and Standley Citation2013), and then the aligned genes were concatenated. Based on the concatenation matrix, a phylogenetic tree was constructed using the maximum likelihood (ML) method implemented in IQ-TREE v. 2.1.2 (Nguyen et al. Citation2015), the best model (TVM + F+R4) was inferred from ModleFinder (Kalyaanamoorthy et al. Citation2017). The bootstrap value was set to 1000. Tree visualization was performed in Figtree v. 1.4.3 (https://github.com/rambaut/figtree/releases).
Results
General features of the chloroplast genome
We analyzed the depth of coverage of the chloroplast genome and tested the annotation accuracy of some difficult genes, the results indicated that the chloroplast genome of V. oxycoccos was trustworthy (Figure S1; Figure S2). The chloroplast genome of V. oxycoccos had a circular tetrameric structure of 177,088 bp in length (, GenBank number: OQ865185), which consisted of a large single copy (LSC) region (104,139 bp), a small single copy (SSC) region (3031 bp), and a pair of inverted repeats (IR) (34,959 bp). This chloroplast genome had a total GC content of 36.74%, the GC content in the IR region (38.49%) was significantly higher than that in the LSC region (35.81%) and the SSC region (28.31%). In addition, the annotation results showed that its chloroplast genome contained 105 different genes, including 73 PCGs, 4 ribosomal RNA genes, and 28 transfer RNA genes (, Table S1). Similar to other Vaccinium species, the four genes (accD, clpP, ycf1, ycf2) were missing, the rps12 gene had only one copy, and some ndh genes appeared in two copies (ndhD, ndhG, ndhE, ndhH, ndhI), and the SSC length was short (Table S1). The SSC region in Vaccinium was more conserved than the LSC and IR regions, which have a high number of variant regions (Figure S3).
Figure 2. The chloroplast genome map of V. oxycoccos. Genes on the inside of the circle are transcribed in a clockwise direction and genes on the outside of the circle are transcribed in a counter-clockwise direction.
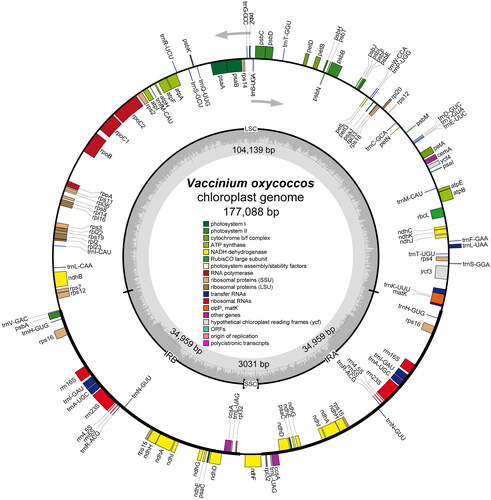
Table 1. Summary of the chloroplast genomes of V. oxycoccos and V. microcarpum species.
Repeat analysis
We identified 143 simple sequence repeats (SSRs) in the chloroplast genome of V. oxycoccos, including 37 mononucleotides, 46 dinucleotides, 15 trinucleotides, 34 tetranucleotides, 6 pentanucleotides, 5 hexanucleotides (). Mononucleotides, dinucleotides and tetranucleotides accounted for most of the SSRs, accounting for 81.82% of the total. The SSRs were highest in the LSC region and lowest in the SSC region, and were mainly concentrated in the non-coding regions (; Table S2). In addition, we identified 50 long repeats, including 24 forward repeats and 26 palindromic repeats (, Table S3), mainly present in the IR region, with a few presents in the LSC region, and none in the SSC region, which is probably related to the short SSC region (Table S3).
IR boundaries analysis
Most Vaccinium species shared some similarity in that the SSC regions were relatively short, the rpl32 and ndhF genes were mainly present near the JSA and JSB boundaries (Figure S4). Compared to closely related species, the overall positions of boundary genes were basically the same, although there was a small amount of contraction and expansion of some genes occurred in the IR region. The IR boundaries of the chloroplast genomes of V. oxycoccos and V. microcarpum were highly similar. Compared with each other, V. oxycoccos showed a small amount of contraction in the IR region.
Phylogenetic analysis
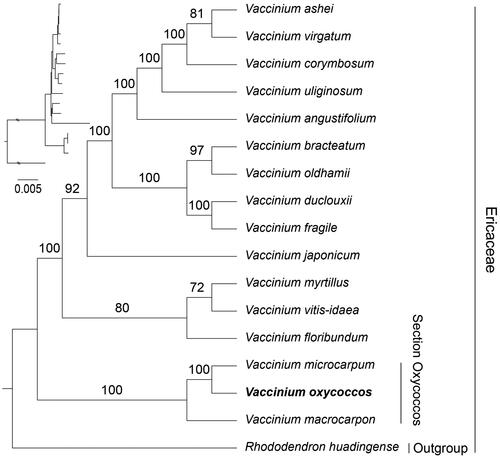
To clarify the phylogenetic position of V. oxycoccos in the Ericaceae, we performed a phylogenetic analysis. The phylogenetic tree showed that our phylogenetic results were generally consistent with previous studies, where most of the nodes have high bootstrap values with high confidence (). The phylogenetic analysis revealed that V. oxycoccos clustered with V. microcarpum and V. macrocarpon, within the section Oxycoccos branch. Moreover, V. oxycoccos was more closely related to V. microcarpum.
Figure 3. Phylogenetic tree based on the concatenated sequences of 74 protein-coding genes in 16 species by maximum-likelihood (ML). values split by backslashes above branches represent ML bootstraps. The best-fit model was TVM + F+R4. Branch supports were tested using ultrafast bootstrap (UFBoot) with 1000 replicates. The scale is 0.005 in the tree. Vaccinium oxycoccos (OQ865185) was marked in bold. The following sequences were used: Vaccinium ashei OP113950, Vaccinium virgatum OM791343 (Fahrenkrog et al. Citation2022), Vaccinium corymbosum OM791342 (Fahrenkrog et al. Citation2022), Vaccinium uliginosum LC521968 (Kim et al. Citation2020), Vaccinium angustifolium OM791344 (Fahrenkrog et al. Citation2022), Vaccinium bracteatum LC521967 (Kim et al. Citation2020), Vaccinium oldhamii MK049537 (Kim et al. Citation2019), Vaccinium duclouxii MK816300 (Chen et al. Citation2019), Vaccinium fragile MK816301 (Guo et al. Citation2019), Vaccinium japonicum MW006668 (Cho et al. Citation2021), Vaccinium myrtillus OM809159 (Fahrenkrog et al. Citation2022), Vaccinium vitis-idaea LC521969 (Kim et al. Citation2020), Vaccinium floribundum OQ331035, Vaccinium microcarpum OQ865186 (Fahrenkrog et al. Citation2022), Vaccinium macrocarpon JQ757046 (Fajardo et al. Citation2013), rhododendron huadingense OM177184 (An et al. Citation2022).
Discussion
Chloroplast genomes are significantly conserved which play a broad role in phylogeny and species identification (Li et al. Citation2021). The chloroplast genomes of Vaccinium species generally had a large total length, a short SSC region, and gene deletions, this phenomenon that requires further in-depth study. The phylogenetic relationship of V. oxycoccos was examined based on the protein-coding genes of the chloroplast genome. Despite revealing the close relationship between V. oxycoccos and V. microcarpum, the phylogenetic analysis was limited by the matrilineal inheritance of the chloroplast genome (Krawczyk et al. Citation2018). An accurate phylogenetic relationship still requires an integrated analysis of the nuclear and organelle genomes (Górniak et al. Citation2010). Due to the complexity of Vaccinium, future genomic analyses of additional Vaccinium species are needed to determine the complex phylogenetic relationships within Vaccinium.
Conclusion
In this study, the chloroplast genome of V. oxycoccos was de novo assembled using short-read data. This chloroplast genome had a typical tetrameric structure similar to that of other Vaccinium species. The chloroplast genome of V. oxycoccos was 177,088 bp in length with a GC content of 36.74%. The LSC, SSC, and IR regions were 104,139 bp, 3031 bp, and 34,959 bp in length, respectively. It contained 105 different genes, including 73 protein-coding genes, 4 rRNA genes, and 28 tRNA genes. Our phylogenetic tree strongly supported the phylogenetic position of V. oxycoccos. These results clearly indicated the close relationship between V. oxycoccos and V. microcarpum. Thus, the chloroplast genome of V. oxycoccos not only enhances the genomic information of Vaccinium but also serves as a foundation for comprehending the evolution of Ericaceae species.
Ethical approval
The Vaccinium oxycoccos is widely cultivated as a fruit plant in China. Experimental researches do not include the genetic transformation, preserving the genetic background of the species used, and any other processes requiring ethics approval. Therefore, no special permission was needed.
Authors’ contributions
Xinrong Qiao was primarily responsible for the design of the experiment; Xinrong Qiao, Qingyi Gu and Run Ye participated in genome assembly and annotation work. Xinrong Qiao, Jing Cai and Nailiang Zhu analyzed and interpreted the data. All authors have read and approved the final manuscript.
Supplemental Material
Download MS Excel (14 KB)Supplemental Material
Download MS Excel (19.2 KB)Supplemental Material
Download MS Excel (12.9 KB)Supplemental Material
Download JPEG Image (1.4 MB)Supplemental Material
Download JPEG Image (2.7 MB)Supplemental Material
Download JPEG Image (318 KB)Supplemental Material
Download JPEG Image (247.1 KB)Disclosure statement
No potential conflict of interest was reported by the authors.
Data availability statement
The genome sequence data that support the findings of this study are available in GenBank of NCBI (http://www.ncbi.nlm.nih.gov/) under the accession no. OQ865185. The associated BioProject, BioSample, and SRA numbers are PRJNA981094, SAMN35661270, SRR24859429, respectively.
Additional information
Funding
References
- Amiryousefi A, Hyvönen J, Poczai P. 2018. IRscope: an online program to visualize the junction sites of chloroplast genomes. Bioinformatics. 34(17):3030–3031. doi: 10.1093/bioinformatics/bty220.
- An R, Niu M, Lou X, Huang H, Lin E., 2022. The complete chloroplast genome of Rhododendron huadingense (Ericaceae). Mitochondrial DNA Part B Resour. 7(11):1910–1912. doi: 10.1080/23802359.2022.2135403.
- Beier S, Thiel T, Münch T, Scholz U, Mascher M., 2017. MISA-web: a web server for microsatellite prediction. Bioinformatics. 33(16):2583–2585. doi: 10.1093/bioinformatics/btx198.
- Chen X, Liu Q, Guo W, Wei H, Wang J, Zhu D, Tan Y., 2019. The complete chloroplast genome of Vaccinium duclouxii, an endemic species in China. Mitochondrial DNA Part B Resour. 4(2):2215–2216. doi: 10.1080/23802359.2019.1624644.
- Cho W-B, Han E-K, Choi I-S, Son DC, Chung GY, Lee J-H., 2021. The complete plastid genome sequence of Vaccinium japonicum (Ericales: Ericaceae), a deciduous broad-leaved shrub endemic to East Asia. Mitochondrial DNA Part B Resour. 6(7):1926–1928. doi: 10.1080/23802359.2021.1935351.
- Daniell H, Jin S, Zhu X-G, Gitzendanner MA, Soltis DE, Soltis PS., 2021. Green giant-a tiny chloroplast genome with mighty power to produce high-value proteins: history and phylogeny. Plant Biotechnol J. 19(3):430–447. doi: 10.1111/pbi.13556.
- Doyle JJ, Doyle JL. 1987. A rapid DNA isolation procedure for small quantities of fresh leaf tissues. Phytochem Bull. 19:11–15.
- Ermis E, Hertel C, Schneider C, Carle R, Stintzing F, Schmidt H., 2015. Characterization of in vitro antifungal activities of small and American cranberry (Vaccinium oxycoccos L. and V. macrocarpon Aiton) and lingonberry (Vaccinium vitis-idaea L.) concentrates in sugar reduced fruit spreads. Int J Food Microbiol. 204:111–117. doi: 10.1016/j.ijfoodmicro.2015.03.017.
- Fahrenkrog AM, Matsumoto GO, Toth K, Jokipii-Lukkari S, Salo HM, Häggman H, Benevenuto J, Munoz PR., 2022. Chloroplast genome assemblies and comparative analyses of commercially important Vaccinium berry crops. Sci Rep. 12(1):21600. doi: 10.1038/s41598-022-25434-5.
- Fajardo D, Senalik D, Ames M, Zhu H, Steffan SA, Harbut R, Polashock J, Vorsa N, Gillespie E, Kron K, et al. 2013. Complete plastid genome sequence of Vaccinium macrocarpon: structure, gene content, and rearrangements revealed by next generation sequencing. Tree Genet Genomes. 9(2):489–498. doi: 10.1007/s11295-012-0573-9.
- Frazer KA, Pachter L, Poliakov A, Rubin EM, Dubchak I., 2004. VISTA: computational tools for comparative genomics. Nucleic Acids Res. 32(Web Server issue):W273–279. doi: 10.1093/nar/gkh458.
- Górniak M, Paun O, Chase MW. 2010. Phylogenetic relationships within Orchidaceae based on a low-copy nuclear coding gene, Xdh: congruence with organellar and nuclear ribosomal DNA results. Mol Phylogenet Evol. 56(2):784–795. doi: 10.1016/j.ympev.2010.03.003.
- Greiner S, Lehwark P, Bock R. 2019. OrganellarGenomeDRAW (OGDRAW) version 1.3.1: expanded toolkit for the graphical visualization of organellar genomes. Nucleic Acids Res. 47(W1):W59–w64. doi: 10.1093/nar/gkz238.
- Guo W, Luo L, Huang Y, Li G, Wang X, Cheng T, Sun Z, Wang F, Zhang L, Li W, et al. 2019. The complete chloroplast genome of Vaccinium fragile (Vacciniaceae), a shrub endemic to China. Mitochondrial DNA Part B Resour. 4(2):2310–2311. doi: 10.1080/23802359.2019.1627948.
- Hancock JF, Lyrene P, Finn CE, et al. 2008. Blueberries and Cranberries. In: Hancock, J.F. (Ed.), Temperate FRUIT crop breeding: Germplasm to genomics. Dordrecht: Springer Netherlands. Chapter 4:115–150.
- Jin J-J, Yu W-B, Yang J-B, Song Y, dePamphilis CW, Yi T-S, Li D-Z., 2020. GetOrganelle: a fast and versatile toolkit for accurate de novo assembly of organelle genomes. Genome Biol. 21(1):241. doi: 10.1186/s13059-020-02154-5.
- Jurikova T, Skrovankova S, Mlcek J, Balla S, Snopek L. 2018. Bioactive compounds, antioxidant activity, and biological effects of European Cranberry (Vaccinium oxycoccos). Molecules. 24(1):24. doi: 10.3390/molecules24010024.
- Kalyaanamoorthy S, Minh BQ, Wong TKF, von Haeseler A, Jermiin LS., 2017. ModelFinder: fast model selection for accurate phylogenetic estimates. Nat Methods. 14(6):587–589. doi: 10.1038/nmeth.4285.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780. doi: 10.1093/molbev/mst010.
- Kim S-C, Baek S-H, Lee J-W, Hyun HJ., 2019. Complete chloroplast genome of Vaccinium oldhamii and phylogenetic analysis. Mitochondrial DNA. Part B Resources. 4(1):902–903. doi: 10.1080/23802359.2019.1579067.
- Kim Y, Shin J, Oh D-R, Kim D-W, Lee H-S, Choi C., 2020. Complete chloroplast genome sequences of Vaccinium bracteatum Thunb., V. vitis-idaea L., and V. uliginosum L. (Ericaceae). Mitochondrial DNA. Part B Resources. 5(2):1843–1844. doi: 10.1080/23802359.2020.1750318.
- Krawczyk K, Nobis M, Myszczyński K, Klichowska E, Sawicki J., 2018. Plastid super-barcodes as a tool for species discrimination in feather grasses (Poaceae: stipa). Sci Rep. 8(1):1924. doi: 10.1038/s41598-018-20399-w.
- Kron KA, Powell EA, Luteyn JL. 2002. Phylogenetic relationships within the blueberry tribe (Vaccinieae, Ericaceae) based on sequence data from MATK and nuclear ribosomal ITS regions, with comments on the placement of Satyria. Am J Bot. 89(2):327–336. doi: 10.3732/ajb.89.2.327.
- Kurtz S, Choudhuri JV, Ohlebusch E, Schleiermacher C, Stoye J, Giegerich R., 2001. REPuter: the manifold applications of repeat analysis on a genomic scale. Nucleic Acids Res. 29(22):4633–4642. doi: 10.1093/nar/29.22.4633.
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R., 2009. The sequence alignment/map format and SAMtools. Bioinformatics. 25(16):2078–2079. doi: 10.1093/bioinformatics/btp352.
- Li J, Tang J, Zeng S, Han F, Yuan J, Yu J., 2021. Comparative plastid genomics of four Pilea (Urticaceae) species: insight into interspecific plastid genome diversity in Pilea. BMC Plant Biol. 21(1):25. doi: 10.1186/s12870-020-02793-7.
- Liu S, Ni Y, Li J, Zhang X, Yang H, Chen H, Liu C., 2023. CPGView: a package for visualizing detailed chloroplast genome structures. Mol Ecol Resour. 23(3):694–704. doi: 10.1111/1755-0998.13729.
- Nguyen L-T, Schmidt HA, von Haeseler A, Minh BQ. 2015. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 32(1):268–274. doi: 10.1093/molbev/msu300.
- Powell EA, Kathleen AK. 2002. Hawaiian blueberries and their relatives—A phylogenetic analysis of Vaccinium sections Macropelma, Myrtillus, and Hemimyrtillus (Ericaceae). Systematic Botany. 27(4):768–779.
- Qu X-J, Moore MJ, Li D-Z, Yi T-S. 2019. PGA: a software package for rapid, accurate, and flexible batch annotation of plastomes. Plant Methods. 15(1):50. doi: 10.1186/s13007-019-0435-7.
- Quinlan AR, Hall IM. 2010. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 26(6):841–842. doi: 10.1093/bioinformatics/btq033.
- Schlautman B, Covarrubias-Pazaran G, Fajardo D, Steffan S, Zalapa J. 2017. Discriminating power of microsatellites in cranberry organelles for taxonomic studies in Vaccinium and Ericaceae. Genet Resour Crop Evol. 64(3):451–466., doi: 10.1007/s10722-016-0371-6.
- Shi L, Chen H, Jiang M, Wang L, Wu X, Huang L, Liu C., 2019. CPGAVAS2, an integrated plastome sequence annotator and analyzer. Nucleic Acids Res. 47(W1):W65–W73. doi: 10.1093/nar/gkz345.
- Sleumer H. 1941. Vaccinioidee-Studien. Botanische Jahrbücher. 71:432–433.
- Tillich M, Lehwark P, Pellizzer T, Ulbricht-Jones ES, Fischer A, Bock R, Greiner S., 2017. GeSeq - versatile and accurate annotation of organelle genomes. Nucleic Acids Res. 45(W1):W6–W11. doi: 10.1093/nar/gkx391.
- Zhang D, Gao F, Jakovlić I, Zou H, Zhang J, Li WX, Wang GT., 2020. PhyloSuite: an integrated and scalable desktop platform for streamlined molecular sequence data management and evolutionary phylogenetics studies. Mol Ecol Resour. 20(1):348–355. doi: 10.1111/1755-0998.13096.
