Abstract
Anaplectoides virens (Noctuoidea: Noctuidae) is a polyphagous herbivorous moth, which feeds on a wide variety of crops. Molecular phylogenetic studies of this species are still limited. We presented the first complete mitochondrial genome of the genus Anaplectoides, which was assembled from data generated using a genome skimming method. The assembled mitogenome is 15,358 bp in length and consists of 37 genes, including 13 protein-coding genes, two rRNAs, 22 tRNAs, and a control region. Except for the start codon of cox1 with CGA, other coding genes use ATN as the start codon. Most PCGs use TAA as the stop codon; however, cox1, cox2, and nad4 use T as the termination codon. Phylogenetic analysis revealed that the genera of ((Agrotis + Striacosta) + Anaplectoides) within Noctuinae formed a monophyletic group. Among Noctuidae, the relationship of ((Noctuinae + Hadninae) + Amphipyrinae) was also highly supported.
Introduction
Noctuidae is the second largest family of Noctuidae in Lepidoptera. Many members in the family are pests that threaten cash crops, reduce production, and destroy the ecological environment (Zhang and Xue Citation2021). Therefore, Noctuidae has received worldwide attention, and researchers are trying to find solutions to control these pests from genome and cell response (Abney et al. Citation2007). Anaplectoides virens Butler, 1878 is considered as one of the most destructive herbivorous pests due to its high reproductive rate. This species is widely distributed in Japan, Korea, and India. Insect mitogenomes are often used for phylogenetic, phylogeographic, and molecular evolutionary analysis because of their unique maternal inheritance, strict homology, and low recombination rate (He et al. Citation2019; Lu et al. Citation2021). To date, the mitogenome of Anaplectoides has not been published. To better facilitate the biological prevention and control of A. virens and the study of phylogenic relationship of subfamilies within Noctuidae, its mitogenome was sequenced and analyzed.
Materials and methods
Female adults of A. virens () were collected in the Sejila Mountain, Linzhi City, Tibet, China (N 29.6241, E 94.6304) in June 2020. The muscle was preserved in 95% ethanol and stored at −20 °C. A specimen was deposited at Insect Specimen Museum of Institute of Plateau Ecology, Tibet Agricultural & Animal Husbandry University (http://www.xza.edu.cn/, Zhaohui Pan, [email protected]) under the voucher number STS-19053.
Figure 1. Species reference image of Anaplectoides virens. The specimen in this photo was collected in Linzhi City of China (coordinates: N 29.6241, E 94.6304). The image was taken by Zhaohui Pan.

Total genomic DNA was extracted from a female adult using an Insect DNA Column Extraction Kit (Baiaolaibo Technology Co., Ltd., Beijing, China). The sample was sequenced by Illumina HiSeq 4000. The raw reads were filtered using trim-galore v0.6.9 (https://anaconda.org/bioconda/trim-galore), quality was assessed using MultiQC v1.12 (https://anaconda.org/bioconda/multiqc). High-quality clean reads were used for the subsequent analysis based on Q20 (≥95%) and Q30 (≥90%). High-quality clean reads were then assembled by SOAPdenovo2 (https://github.com/aquaskyline/SOAPdenovo2), with the mitogenome of Agrotis ipsilon Hufnagel, 1766 used as the reference (GenBank accession number: NC022185) (Wu et al. Citation2015). The mitogenome was annotated by the MITOS webserver (Bernt et al. Citation2013) and visualized by CGView Web server (https://cgview.ca/). To determine the correctness of assembly, coverage depth was calculated by mapping reads onto the mitogenome sequence using bowtie2 v2.3.4.3 (Langmead and Salzberg Citation2012).
Phylogenetic analysis was performed on concatenated nucleotide sequences of 13 protein-coding genes (PCGs) derived from eight Noctuidae species, with two Sphingidae and one species as the outgroups. Nucleotide sequences for each of the 13 PCGs were translated into amino acids, aligned separately with MUSCLE implemented within MEGA v6.05 (https://www.megasoftware.net/), and then toggled back into nucleotide alignments. Gblock v0.91b (Castresana Citation2000) was also employed to eliminate the poorly aligned position and divergent regions. Phylogenetic trees were constructed using PhyloSuite v1.2.2 (Zhang et al. Citation2020). The topological consistency between two trees was calculated using Consel v0.1i (Shimodaira and Hasegawa Citation2001), and the optimal topology was selected and displayed.
Results
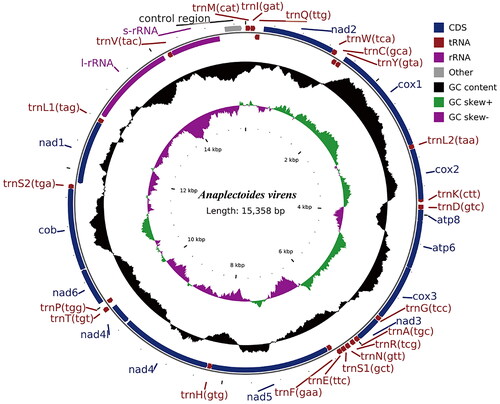
The complete mitogenome of A. virens is a circular molecule of 15,358 bp in length (GenBank accession number: MZ707539). It contains 13 PCGs, 22 transfer RNAs (tRNAs), two ribosomal RNA (rRNA) genes, and a noncoding control region (D-loop) (). Except for cox1, all other coding genes use ATN as the start codon. Most PCGs use TAA as the stop codon; however, cox1, cox2, and nad4 use T as the termination codon. The length of the tRNA genes ranged from 64 to 72 bp, with a total length of 1468 bp. The lengths of the 16S and 12S were 1356 and 786 bp, respectively. The mitogenome of A. virens was assembled according to depth of the coverage (high coverage of over 26×) (Figure S1).
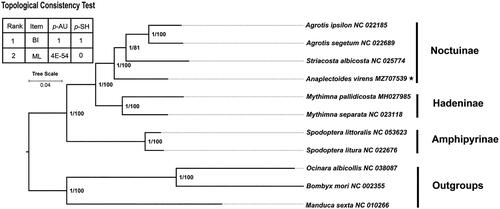
Phylogenetic results indicated that the three subfamilies were a monophyletic group. The phylogenetic relationships among subfamilies are as follows: (Amphipyrinae + (Hadeninae + Noctuinae)) (). In this study, A. virens is located at the base of the subfamily Noctuinae, with the phylogenetic relationship of (A. virens + (Striacosta albicosta + (A. ipsilon + Agrotis segetum))).
Figure 3. Phylogenetic tree was constructed based on 13 PCGs of eight Noctuidae mitogenomes and three outgroups. The support values of the corresponding nodes are shown (bootstrap for BI on the left and posterior probability for ML on the right). The result of the topological consistency test is shown in the upper left corner. AU: approximate unbiased test; SH: Shimodaira-Hasegawa test.
Discussion and conclusions
In this study, we first reported the mitogenome of A. virens. Phylogeny inferred using the ML and BI method based on 13 PCGs showed that Noctuinae was the sister lineage of Hadeninae, which was consistent with the result based on morphological (Speidel et al. Citation1996) and molecular phylogeny studies (Li et al. Citation2021). In this study, A. virens was located at the base of the subfamily Noctuinae, with the phylogenetic relationship of (A. virens + (S. albicosta + (A. ipsilon + A. segetum))). This study not only provided important molecular data for the evolutionary and phylogeographic analysis of A. virens, but also provided a basis for further study of the phylogenetic relationships within Chalcosiinae cutworm moths.
Author contributions
WZ conceived the study; ZP collected the samples and performed the experiments; ZP and SH analyzed the data and prepared the manuscript; WZ revised the manuscript. All authors agree to be accountable for all aspects of the work.
Supplemental Material
Download MS Word (182.9 KB)Acknowledgements
Thanks to Dunyuan Huang for submitting the data to NCBI.
Disclosure statement
No potential conflict of interest was reported by the authors.
Data availability statement
The mitogenome sequence data that support the findings of this study are openly available in GenBank of NCBI at https://www.ncbi.nlm.nih.gov/ under the accession no. MZ707539. The original data were submitted in NCBI, BioProject: PRJNA826705, SRA: SRR18750010, and BioSample: SAMN27578231.
Additional information
Funding
References
- Abney MR, Sorenson CE, Southern PS. 2007. Pyrethroid insecticide efficacy against tobacco budworm (Lepidoptera: Noctuidae) in North Carolina flue-cured tobacco: implications for insecticide resistance management. J Entomol Sci. 42(4):582–588. doi: 10.18474/0749-8004-42.4.582.
- Bernt M, Donath A, Jühling F, Externbrink F, Florentz C, Fritzsch G, Pütz J, Middendorf M, Stadler PF. 2013. MITOS: improved de novo metazoan mitochondrial genome annotation. Mol Phylogenet Evol. 69(2):313–319. doi: 10.1016/j.ympev.2012.08.023.
- Castresana J. 2000. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol Biol Evol. 17(4):540–552. doi: 10.1093/oxfordjournals.molbev.a026334.
- He B, Su TJ, Niu ZQ, Zhou ZY, Gu ZY, Huang DY. 2019. Characterization of mitogenomes of three Andrena bees (Apoidea: Andrenidae) and insights into the phylogenetics. Int J Biol Macromol. 127:118–125. doi: 10.1016/j.ijbiomac.2019.01.036.
- Langmead B, Salzberg SL. 2012. Fast gapped-read alignment with Bowtie 2. Nat Methods. 9(4):357–359. doi: 10.1038/nmeth.1923.
- Li Z, Wang W, Zhang L. 2021. Complete mitochondrial genome of Spodoptera littoralis (Lepidoptera: Noctuidae) from Egypt. Mitochondrial DNA B. 6(2):432–434. doi: 10.1080/23802359.2020.1870894.
- Lu H, He B, Hao Y, Zhou Z, Su C, Huang D. 2021. Comparative mitogenomic analysis of two cuckoo bees (Apoidea: Anthophila: Megachilidae) with phylogenetic implications. Insects. 12(1):29. doi: 10.3390/insects12010029.
- Shimodaira H, Hasegawa M. 2001. Consel: for assessing the confidence of phylogenetic tree selection. Bioinformatics. 17(12):1246–1247. doi: 10.1093/bioinformatics/17.12.1246.
- Speidel W, Fänger H, Naumann CM. 1996. The phylogeny of the Noctuidae (Lepidoptera). Syst Entomol. 21(3):219–252. doi: 10.1046/j.1365-3113.1996.d01-14.x.
- Wu QL, Cui WX, Wei SJ. 2015. Characterization of the complete mitochondrial genome of the black cutworm Agrotis ipsilon (Lepidoptera: Noctuidae). Mitochondrial DNA. 26(1):139–140. doi: 10.3109/19401736.2013.815175.
- Zhang D, Gao F, Jakovlić I, Zou H, Zhang J, Li WX, Wang GT. 2020. PhyloSuite: an integrated and scalable desktop platform for streamlined molecular sequence data management and evolutionary phylogenetics studies. Mol Ecol Resour. 20(1):348–355. doi: 10.1111/1755-0998.13096.
- Zhang Y, Xue S. 2021. Characterization and phylogenetic analysis of the mitochondrial genome of Macdunnoughia hybrida (Lepidoptera: Noctuidae: Plusiinae). Mitochondrial DNA B Resour. 6(8):2294–2296. doi: 10.1080/23802359.2021.1945972.