Abstract
In this study, we assembled the complete chloroplast (cp) genome of Cynanchum acutum subsp. sibiricum using high-throughput Illumina sequencing reads. The resulting chloroplast genome assembly displayed a typical quadripartite structure with a total length of 158,283 bp, which contained a pair of inverted repeat regions (IRs) of 24,459 bp. These two IRs were separated by a large single-copy region (LSC) and a small single-copy region (SSC) of 89,424 bp and 19,941 bp in length, respectively. The C. acutum subsp. sibiricum cp genome contained 130 genes, and its overall GC content was 37.87%. Phylogenetic analysis among C. acutum subsp. sibiricum and nine other Cynanchum species demonstrated that C. acutum subsp. sibiricum was closely related to C. chinense. The C. acutum subsp. sibiricum cp genome presented in this study lays a good foundation for further genetic and genomic studies of the Cynanchum as well as Apocynaceae.
1. Introduction
Cynanchum acutum L. subsp. sibiricum (Willd.) K. H. Rech 1970 (Rechinger, Citation1970), a medicinal plant, belongs to Apocynaceae (Li et al. Citation1995). It is distributed in the northwest of China, including Inner Mongolia, Ningxia, Gansu and Xinjiang (Li et al. 1995). The dried whole plant of C. acutum subsp. sibiricum was used as a traditional Chinese medicine to treat bleeding and inflammation (Liu et al. Citation2007). This medicinal plant has been used for the therapy of chronic tracheitis in China (Yildiz et al. Citation2017).
Recent usage of the complete chloroplast genome in solving phylogenetic problem has been on the rise due to its unique nature and its capability to unravel valuable information worthy of adopting (Abba et al. Citation2021). Complete chloroplast genomes have been used vividly in resolving some unanswered questions in plant taxonomy recently (Ruhsam et al. Citation2015). In addition to the evolutionary perspective, the chloroplast genome has important implications in chloroplast transformation (Daniell et al. Citation2008). A systematic study of C. acutum subsp. sibiricum would have significant implications for understanding the origin and evolution of the Cynanchum and local flora. However, the chloroplast genome of C. acutum subsp. sibiricum has not been reported.
In this study, the complete chloroplast genome of C. acutum subsp. sibiricum has been assembled in order to lay a foundation for further research.
2. Materials
Fresh leaves of C. acutum subsp. sibiricum were collected from Pingluo (Shizuishan, Ningxia, China; coordinates: 106.3849E, 38.8320 N) (by Lei Zhang: [email protected]) (), and dried with silica gel. The voucher specimen was stored in Herbarium of North Minzu University with an accession number of zlnmu2021151 ().
3. Methods
1.0 µg of the leaf sample was measured for DNA preparation. Total genomic DNA was extracted with a modified CTAB method (Doyle and Doyle Citation1987). NEBNext DNA Library Kit was used to generate the sequencing libraries following the manufacturer’s manual. DNA was fragmented erratically to 350 bp size by shearings. This library was sequenced on the Illumina NovaSeq 6000 platform with 150 bp paired-end read length. We obtained 6.2 Gb high quality pair-end reads for C. acutum subsp. sibiricum. After removing the adapters, we de novo assembled the cp genome of C. acutum subsp. sibiricum using NOVOPlasty 4.1 (Dierckxsens et al. Citation2017) using the following parameters: k-mer = 39 and genome range 120,000–200,000 bp. The complete chloroplast genome sequence of C. auriculatum (KU900231) was used as a reference. Plann v1.1 (Huang and Cronk Citation2015) was used to annotate the chloroplast genome and Geneious v11.0.3 (Kearse et al. Citation2012) was employed to correct the annotation. In order to further clarify the phylogenetic position of C. acutum subsp. sibiricum, chloroplast genome of nine representative species were obtained from NCBI GenBank to reconstruct the chloroplast genome phylogenetic tree, with Nerium oleander being used as an outgroup. All the sequences were aligned using MAFFT v.7.313 (Katoh and Standley Citation2013) and maximum likelihood phylogenetic analyses were conducted by using RAxML v.8.2.11 (Stamatakis Citation2014) under GTRCAT model with 1000 bootstrap replicates.
4. Results
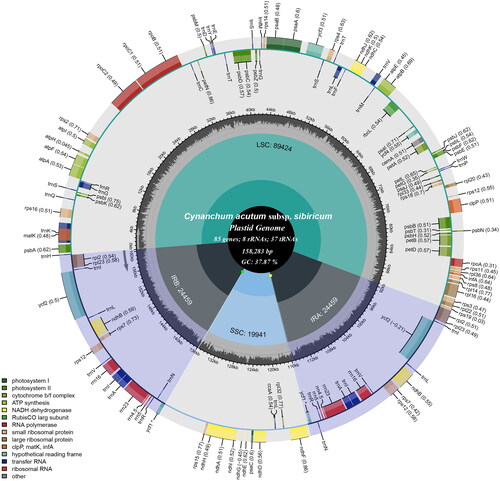
The total chloroplast genome of C. acutum subsp. sibiricum (OQ390041) was 158,283 bp in length and depth for average, maximal and minimal were 7893.73 x, 8022 x and 97 x (Supplementary Figure 1), with a typical quadripartite structural organization, consisting of a large single copy (LSC) region of 89,424 bp, two inverted repeat (IR) regions of 24,459 bp and a small single copy (SSC) region of 19,941 bp (). The cp genome contained 130 complete genes, including 85 protein-coding genes (85 PCGs), 8 ribosomal RNA genes (8 rRNAs), and 37 tRNA genes (37 tRNAs). Most genes were occurred in a single copy, while 16 genes occurred in double, including four rRNAs (rrn16, rrn23, rrn4.5 and rrn5), four tRNAs (trnA-UGC, trnI-CAU, trnI-GAU, trnL-CAA, trnN-GUU, trnR-ACG, and trnV-GAC), and 5 PCGs (rpl23, rps7, ndhB, ycf2 and rpl2). Meanwhile, The cp genome contained 13 cis-splicing genes (Supplementary Figure 2) and 1 trans-splicing gene (Supplementary Figure 3). The overall GC content of cp DNA is 37.87%, the respective values for the LSC, SSC, and IR regions were 36.15%, 32.00%, and 43.43%.
Figure 2. The detailed genome map of Cynanchum acutum subsp. sibiricum cp genome. The large single copy (LSC), small single copy (SSC) region and two inverted repeat regions (IRA and IRB), and GC content (light gray) are shown in the inside track. Gene models including protein-coding genes, tRNA genes, and rRNA genes are shown with various colored boxes in the outer track.
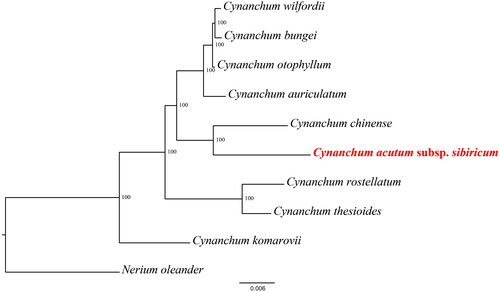
The phylogenetic tree showed that the species of Cynanchum were divided into three subclades (). C. thesioides and C. rostellatum clustered the second subclade, and the C. chinense and C. acutum subsp. sibiricum into another subclade. In addition, C. acutum subsp. sibiricum has the closest relationship with C. chinense.
Figure 3. Phylogenetic relationships of Cynanchum species using whole chloroplast genome. GenBank accession numbers: Cynanchum acutum subsp. sibiricum OQ390041, Cynanchum auriculatum KU900231 (Yu et al. Citation2021), Cynanchum bungei OK271106 (Pei et al. Citation2022), Cynanchum chinense MW415427 (Chen and Zhang Citation2022), Cynanchum otophyllum OQ587923, Cynanchum komarovii ON854661(Liu et al. Citation2023), Cynanchum thesioides MW864598 (Kang et al. Citation2021), Cynanchum rostellatum OQ852689 (Lee et al. Citation2022), Cynanchum wilfordii KX352467 (Hyun-Seung, Citation2016) and Nerium oleander KJ953907.
5. Discussion and conclusion
The chloroplast genome of angiosperms provides a powerful tool for species tree estimation, population genetic analysis, chloroplast gene function and chloroplast metabolism analysis, and investigation of species adaptation to extreme environments (Mehmood et al. Citation2020 a,b,c). Here, we reported the first chloroplast genome for C. acutum subsp. sibiricum. This cp genome will provide useful genetic resource for further genetic and genomic studies of the genus Cynanchum and Apocynaceae.
The phylogenetic tree showed that the species of Cynanchum were divided into three subclades. All 8 species and varieties of the ingroup, except C. komarovii, comprise a group that supports the monophyly of the Cynanchum clade, This phylogenetic result was similar to those of Lee et al. (Citation2022) and Pei et al. (Citation2022). Further studies of the C. komarovii is in order, especially to confirm the relation-ships of this species to Cynanchum. Increasing the number of cp genomes of Cynanchum and Apocynaceae will provide deeper insights into the evolutionary of this ecologically and economically important family.
Author contributions
Lei Zhang conceived and supervised the project. Erdong Zhang, Yi Liu, Yan Wang and Xuedan Zhang analyzed the data and wrote the manuscript. All of the authors have read and approved the final manuscript and have agreed to be accountable for all aspects of the work.
Supplemental Material
Download MS Word (1.2 MB)Acknowledgments
The authors are really grateful for the access to raw genome data from public database.
Disclosure statement
The coauthors do not have any conflict of interest to declare. The authors alone are responsible for the content and composition of the paper. Additionally, fresh and mature leaves from an adult plant of C. acutum subsp. sibiricum were taken with the permission of the community management department.
Data availability statement
The sequenced data supporting the findings of this study are openly available in NCBI (https://www.ncbi.nlm.nih.gov/) under the accession no. OQ390041. The associated BioProject, SRA, and Bio-Sample numbers are PRJNA930674, SRR23341524, and SAMN33018279, respectively.
Additional information
Funding
References
- Abba A, Alzahrani D, Yaradua S, Albokhari E. 2021. Comparative chloroplast genome analysis and evolutionary relationships in some species of Asclepiadeae, Apocynaceae. 0: 1–7. doi: 10.18805/LR-565.
- Chen G, Zhang X. 2022. The complete chloroplast genome of Chinese medicinal herb Cynanchum chinense R. Br. (Apocynaceae) and its phylogenetic position. Mitochondrial DNA Part B Resour. 7(4):598–599. doi: 10.1080/23802359.2021.1959434.
- Daniell H, Wurdack JK, Kanagaraj A, Lee SB, Saski C, Jansen RK. 2008. The complete nucleotide sequence of the cassava (Manihot esculenta) chloroplast genome and the evolution of atpF in Malpighiales: RNA editing and multiple losses of a group II intron. Theor Appl Genet. 116(5):723–737.
- Dierckxsens N, Mardulyn P, Smits G. 2017. NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 45(4):e18. doi: 10.1093/nar/gkw955.
- Doyle JJ, Doyle JL. 1987. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull. 19:11–15.
- Huang D, Cronk Q. 2015. Plann: a command-line application for anno-tating plastome sequences. Appl Plant Sci. 3(8):1500026. doi: 10.3732/apps.1500026.
- Hyun-Seung P, Kyu-Yeob Kim, Lee K, et al. 2016. The complete chloroplast genome sequence of an important medicinal plant Cynanchum wilfordii (Maxim.) Hemsl. (Apocynaceae). Mitochondrial DNA. Part A. 27:3747–3748.
- Kang P, Guo Y, Zhang Y, Wei Y., 2021. The complete chloroplast genome sequence of medicinal plant: cynanchum thesioides (Asclepiadaceae). Mitochondrial DNA Part B Resour. 6(9):2592–2593. doi: 10.1080/23802359.2021.1961622.
- Katoh K, Standley D. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780. doi: 10.1093/molbev/mst010.
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, et al. 2012. Geneious basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 28(12):1647–1649. doi: 10.1093/bioinformatics/bts199.
- Lee SH, Jang W, Kim E, Kim J, Gong H, Kang J-S, Shim H, Park JY, Yang T-J., 2022. The complete plastome of Cynanchum rostellatum (Apocynaceae), an indigenous plant in Korea. Mitochondrial DNA Part B Resour. 7(12):2035–2039. doi: 10.1080/23802359.2022.2148489.
- Li PT, Michael GG, Stevens WD. 1995. Asclepiadaceae. In: Wu Z, Raven P (editors). Flora of China. 16:311–313. Beijing, China: Science Press.
- Liu W, Wang Z, Tian Y, Ji B. 2023. The complete chloroplast genome sequence of Vincetoxicum mongolicum (Apocynaceae), a perennial medicinal herb. Genet Mol Biol. 46(2):e20220303., doi: 10.1590/1678-4685-GMB-2022-0303.
- Mehmood F, Ubaid Z, Shahzadi I, Ahmed I, Waheed, M. T, Poczai P, Mirza B., Abdullah , (2020a). Plastid genomics of Nicotiana (Solanaceae): insights into molecular evolution, positive selection and the origin of the maternal genome of Aztec tobacco (Nicotiana rustica). PeerJ. 8: e9552. doi: 10.7717/peerj.9552.
- Mehmood F, Ubaid Z, Bao Y, Poczai P, Mirza B, Abdullah , (2020b) Comparative plastomics of Ashwagandha (Withania, Solanaceae) and identification of mutational hotspots for barcoding medicinal plants. Plants. 9(6): 752. doi: 10.3390/plants9060752.
- Mehmood, F, Shahzadi I, Ahmed I, Waheed M. T., Mirza B, Abdullah , (2020c) Characterization of Withania somnifera chloroplast genome and its comparison with other selected species of Solanaceae. Genomics. 112(2): 1522–1530. doi: 10.1016/j.ygeno.2019.08.024.
- Pei L, Shu S, Ji B, Cui N., 2022. Complete sequence of Cynanchum rostellatum (Apocynaceae: Asclepiadoideae) chloroplast genome and its phylogenetic analysis. Mitochondrial DNA Part B Resour. 7(7):1395–1397. doi: 10.1080/23802359.2022.2102444.
- Rechinger KH. 1970. Flora des Iranischen Hochlandes und der Umrahmenden Gebirge : Persien, Afghanistan, Teile von West-Pakistan, Nord-Iraq, (cont). Flora Iranica. 73:9.
- Ruhsam M, Rai HS, Mathews S, Ross TG, Graham SW, Raubeson LA, Mei W, Thomas PI, Gardner MF, Ennos RA, et al. 2015. Doescomplete plastid genome sequencing improve species discrimination and phylogenetic resolution in Araucaria? Mol Ecol Resour. 15(5):1067–1078. doi: 10.1111/1755-0998.12375.
- Yildiz I, Sen O, Erenler R, Demirtas I, Behcet L. 2017. Behcet bioactivity-guided isolation of flavonoids from Cynanchum acutum L. subsp. sibiricum (willd.) Rech. f. and investigation of their antiproliferative activity. Nat Prod Res. 2017:1–5.
- Yu X, Wang W, Yang H, Zhang X, Wang D, Tian X., 2021. Transcriptome and comparative chloroplast genome analysis of Vincetoxicum versicolor: insights into molecular evolution and phylogenetic implication. Front Genet. 12:602528. doi: 10.3389/fgene.2021.602528.
- Liu Y, Qu J, Yu SS, Hu YC, Huang XZ. 2007. Seven new steroidal glycosides from the roots of Cynanchum forrestii. Steroids. 72(4):313–322. doi: 10.1016/j.steroids.2006.11.024.
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30(9):1312–1313. doi: 10.1093/bioinformatics/btu033.

