Abstract
The complete mitochondrial genome of Pachycephus smyrnensis Stein, 1876 collected from Sivas, Turkey, is described. The circled genome is 20,393 bp in length and contains a typical set of 37 genes. The missing control regions, trnQ and trnI in previously reported P. smyrnensis (KX907846) were obtained in this precise assembly based on a considerable amount of raw data. A denser sampled phylogenetic analysis shows that the two P. smyrnensis constitute a branch sister to P. cruentatus (Eversmann, Citation1847). Pachycephus is a sister group of Phylloecus within Hartigiinae and remote from Characopygus, a genus within Cephinae. The monophyly of Pachycephini has been rejected.
Introduction
The natural systematic frame of Cephidae has not been resolved till present. Liu et al. (Citation2022) just revealed the paraphyly of Syrista and reestablished the genus Neosyrista Benson. The validity of Pachycephini as well as its systematic position within Cephidae is also one of the remaining problems. Benson (Citation1946) first proposed the tribe Pachycephini under the subfamily Cephinae, and two genera were placed into the tribe: Pachycephus Stein and Characopygus Konow. This system was followed by Muche (Citation1981) and Abe and Smith (Citation1991). The three diagnostic characters of Pachycephini proposed by Benson (Citation1946) were: the ‘face’ is very broad with a distance between the two anterior tentorial pits about two times the distance between a torulus and an anterior tentorial pit at same side; the left mandible stout with a long outer tooth and two short inner teeth of almost equal prominence; and the male apical sternite constricted before apex where remains a small apical lobe. In the past more than 70 years, there was no study debating the validity and the systematic position of Pachycephini within Cephidae. Recently Liu (Citation2019) studied the morphology and classification of Hartigiinae from China and found that the broad ‘face’ of Cephidae was probably a plesiomorphic character, and the character state of mandibles and male genitalia are varied greatly among different genera of Cephidae. So the basis of the monophyly of Pachycephini is uncertain. Based on COI data, Zhang (Citation2020) analyzed the phylogeny of Cephidae and also debated the monophyly of Pachycephini. She found that Pachycephus Stein may be a member of Hartigiinae and it is not a sister group of Characopygus Konow.
In this study, we sequenced the mitochondrial genome (mt genome) of Pachycephus smyrnensis Stein, 1876 (OL077523) and inferred the phylogeny of Cephidae with all previously reported mt genomes of Cephidae to clarify the phylogenetic position of Pachycephus within the Cephidae and the monophyly of the tribe Pachycephini.
Materials and methods
A voucher specimen (CSCS-Hym-MC0233) was deposited in the Asia Sawfly Museum, Nanchang (ASMN). It was collected from Sivas Cumhuriyet University Campus, Sivas, Turkey (39.70 N, 37.02 E) in June 2018. The specimen was identified by Wei Meicai ([email protected]), namely Pachycephus smyrnensis Stein, 1876. And the reference specimen was exchanged with DEI and is kept in ASMN (). Whole genomic DNA was extracted from the thorax muscle of the female (SAMN22566689) using the DNeasyR Blood & Tissue Kits (Qiagen, Valencia, CA). Genomic DNA was sequenced by the high-throughput Illumina Hiseq 4000 platform, yielding a total of 328,645,942 raw reads (SRR16627925). DNA sequences were assembled by MitoZ (Meng et al. Citation2019) and NOVOPlasty (Dierckxsens et al. Citation2017), and further verified by Geneious Prime 2019.2.1 (Biomatters, Auckland, New Zealand, https://www.geneious.com). A read coverage plot for studies using next-generation DNA sequencing technologies was provided in the supplementary material. The sequences were multiply aligned using MAFFT method in the TranslatorX server (Abascal et al. Citation2010). Annotations were first generated in MITOS web server (Bernt et al. Citation2013), then corrected in Geneious if necessary. The phylogenetic tree inferred Bayesian inference using MrBayes version 3.2.6 (Ronquist et al. Citation2012).
Results
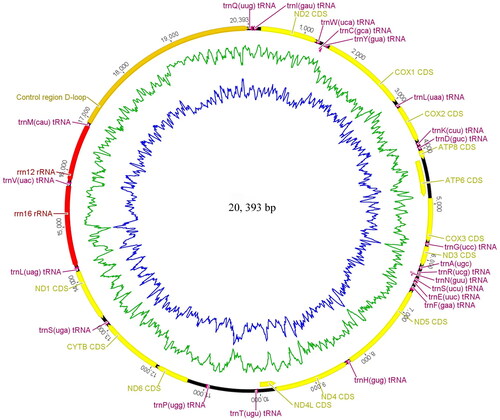
The sequence yield by MitoZ was 18,442 bp in length, and yield by NOVOPlasty was 20,750 bp in length. Two results have a completely identical region, containing 37 genes and an incomplete control region (CR), which was further examined by reassembly using Stenocephus shenyang (unpublished) and Phylloecus fuscicosta (unpublished) as references (coverage was 102, 746, and 104, 212, respectively). A 124 bp overlap was found in the latter two sequences, which can guide the closure of the genome. Then, we used the whole non-coding region itself as a reference to examine the longest interval region further. By consistently obtaining similar coverage of the assembly contigs, we were able to confirm the 3467 bp control region between trnQ and trnM. And we obtained the complete mitochondrial genome finally. The mt genome circular () was maked using Geneious Prime under the circular sequence option.
Figure 3. Phylogenetic tree including Pachycephus smyrnensis based on the combination of 13 nucleotide sequences. Numbers at the left of nodes are bootstrap support values. The accession number of each species is indicated after the Latin name.

The length of the complete mt genome of P. smyrnensis is 20,393 bp, and it contains 37 genes, including 13 protein-coding genes (PCGs), 22 transfer RNA genes (tRNAs), two ribosomal RNA genes (rRNAs), and a 3467-bp CR (). The A + T content of P. smyrnensis mt genome is 80.8% (A: 40.8%, T: 40.0%, C: 12.5%, G: 6.6%). All PCGs initiated by ATN codons and most PCGs ended with TAA stop codons, except nad5 using TAG as a stop codon. Six tRNAs (trnQ, trnW, trnF, trnH, trnP, and trnV), two rRNAs, and four PCGs (nad1, nad4, nad4L, and nad5), were located on the N-strand, while the J-strand encoded the remaining.
Phylogenetic analysis showed the two P. smyrnensis formed a clade which sister to P. cruentatus (Eversmann, Citation1847). The monophyletic Pachycephus is a sister group of Phylloecus within Hartigiinae and it is remote from Characopygus, a genus within Cephinae and standing between Cephus and Calameuta. The result also revealed that the subfamily Cephinae composed of Cephus, Calameuta and Characopygus is a monophyletic group, and the genus Janus is probably a paraphyletic group.
Discussion
The genome architecture is almost consistent with the previously reported P. smyrnensis (KX907846) which lacks trnQ, trnI, and control region. The features of base composition and codon usage are highly similar. A total of 75 single base substitutions in 13 PCGs were found. In addition, by comparing with the other species of Cephidae, we detected a 508 bp long sequence which was repeated three times in the CR. This is consistent with the characteristics of the Cephidae mt genome.
The monophyly of Pachycephini was rejected. However, more Cephidae material and mt genome data are needed to gain a high-resolution phylogeny of Cephidae for clarifying detailed phylogeny of Hartigiinae in the future.
Ethics statement
The collection of specimens conformed to the requirement of International ethics, which are unrestricted species. The process and purpose of this experimental research were in line with the rules and regulations of our institute. There are no ethical issues and other conflicts of interest in this study.
Author contributions
Mengmeng Liu: formal analysis, writing-original draft, writing-review and editing, funding acquisition. Min Xu: formal analysis, writing-original draft, software. Zejian Li and Meicai Wei: conceptualization, methodology, resources, supervision, funding acquisition. Mahir Budak: resources, formal analysis, writing-original draft. Lin Liu: data curation, validation, visualization. All authors have read and agreed to the published version of the manuscript.
Supplemental Material
Download TIFF Image (9.1 MB)Acknowledgments
We thank the Lab of Insect Systematics and Evolutionary Biology (LISEB) from Jiangxi Normal University, notable Niu gengyun, for her kindly assistant on data curation. The valuable comments of the anonymous reviewers are gratefully acknowledged.
Disclosure statement
No potential conflict of interest was reported by the authors.
Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI at [https://www.ncbi.nlm.nih.gov] (https://www.ncbi.nlm.nih.gov/) under the accession number OL077523. The associated BioProject, SRA, and BioSample numbers are PRJNA774478, SRR16627925, and SAMN22566689, respectively. All related files had been uploaded to figshare https://figshare.com/projects/Pachycephus_smyrnensis/127397.
Additional information
Funding
References
- Abascal F, Zardoya R, Telford MJ. 2010. TranslatorX: multiple alignment of nucleotide sequences guided by amino acid translations. Nucleic Acids Res. 38:W7–13. doi: 10.1093/nar/gkq291.
- Abe M, Smith DR. 1991. The genus-group names of Symphyta (Hymenoptera) and their type species. Esakia, Fukuoka. 31:1–115. doi: 10.5109/2551.
- Benson RB. 1946. Classification of the Cephidae (Hymenoptera Symphyta). Trans R Entomol Soc Lond. 96(6):89–108. doi: 10.1111/j.1365-2311.1946.tb00445.x.
- Bernt M, Donath A, Jühling F, Externbrink F, Florentz C, Fritzsch G, Pütz J, Middendorf M, Stadler PF. 2013. MITOS: improved de novo metazoan mitochondrial genome annotation. Mol Phylogenet Evol. 69(2):313–319. doi: 10.1016/j.ympev.2012.08.023.
- Dierckxsens N, Mardulyn P, Smits G. 2017. NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 45(4):e18. doi: 10.1093/nar/gkw955.
- Eversmann E. 1847. Fauna hymenopterologica volgo-uralensis exhibens Hymenopterorum species quas in provinciis Volgam fluvium inter et montes Uralenses sitis observavit et nunc descripsit. Bulletin de la Société Impériale des Naturalistes de Moscou. 20(1): 3–68.
- Liu L. 2019. A systematic study of Hartigiinae (Hymenoptera: Cephidae) [master thesis]. Changsha, China: Central South University of Forestry and Technology; p. 1–157.
- Liu MM, Sun ZM, Özgül D, Wei MC, Liu L. 2022. The complete mitochondrial genome of Syrista parreysii (Spinola, 1843) (Hymenoptera: Cephidae) and phylogenetic analysis. Mitochondrial DNA B Resour. 7(9):1737–1739. doi: 10.1080/23802359.2022.2124828.
- Meng G, Li Y, Yang C, Liu S. 2019. MitoZ: a toolkit for animal mitochondrial genome assembly, annotation and visualization. Nucleic Acids Res. 47(11):e63. doi: 10.1093/nar/gkz173.
- Muche H. 1981. Die Cephidae der Erde (Hym., Cephidae). Dtsch Entomol Z. 28(4–5):239–295. doi: 10.1002/mmnd.19810280405.
- Ronquist F, Teslenko M, Van Der Mark P, Ayres DL, Darling A, H€ohna S, Larget B, Liu L, Suchard MA, Huelsenbeck JP. 2012. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol. 61(3):539–542. doi: 10.1093/sysbio/sys029.
- Zhang Y. 2020. Study on the phylogeny of Cephidae based on mitochondrial rRNA data [master thesis]. Changsha, China: Central South University of Forestry and Technology. p. 1–103.