Abstract
The complete mitochondrial genome of Pyricularia oryzae Cavara 1892 strain Guy11 is 34,865 bp in length (GenBank accession number OP095391), containing 29 tRNA genes, 2 rRNA genes, and 15 protein-coding genes (PCGs). The gene order and orientation are novel compared to other Sordariomycetes species with sequenced mitogenomes in the GenBank database. Phylogenetic analysis suggests that P. oryzae Guy11 and 19 other Sordariomycetes species form a monophyletic group. The complete mitochondrial sequence of P. oryzae Guy11 will be a valuable resource for species identification, population genetics, phylogenetics, and comparative genomics studies in Sordariomycetes and Magnaporthales.
Introduction
Pyricularia oryzae (sexual state: Magnaporthe oryzae), the causal agent of the rice blast disease, is a well-known pathogenic fungus in worldwide agriculture. Outbreaks of rice blast constitute a significant constraint to world cereal production and food security. It is estimated that the annual yield loss of rice is sufficient to feed more than 60 million people (Nalley et al. Citation2016). The resistant rice cultivars will lose their resistance upon planting for 3–5 years as P. oryzae evolves rapidly. The fungus causes blast disease on a vast range of economically important species, including rice, wheat, barley, millet, maize (Pordel et al. Citation2021), and other species of the Poaceae (Gramineae). It is now known that P. grisea consists of a cryptic species complex containing at least two groups based on distinct genetic differences and a lack of interbreeding capacity (Zhang et al. Citation2016). Strains isolated from Digitaria have been defined as P. grisea, whereas isolates from rice and other hosts have been named P. oryzae (Klaubauf et al. Citation2014). The current P. oryzae reference genome (MG8) is based on the laboratory strain 70–15 (Dean et al. Citation2005), which is a progeny of the intercross between strain CH104-3 (rice isolate) and AR-4 (weeping love grass isolate), with the backcross parent of Guy11 (rice isolate) (Chao and Ellingboe Citation1991). The mitochondrial sequence is still absent from the reference genome (https://fungi.ensembl.org/Magnaporthe_oryzae/Info/Index). Therefore, in this study, the complete mitochondrial sequence of P. oryzae Guy11 was assembled and compared to other Sordariomycetes species. This result could be helpful for species identification and the genetics and evolutionary processes of P. oryzae and other Sordariomycetes species.
Materials
Pyricularia oryzae Cavara 1892 strain Guy11 (ATCC® 201236™) was first isolated in 1979 from diseased rice leaves collected by Notteghem in French Guiana (Leung et al. Citation1988). The original strain from ATCC in 2013 was maintained and asexually propagated at Xiaohong’s lab at Zhejiang University. Specimen for sequencing was collected from Guy11 cultures grown in liquid medium by Xiaohong in the lab in Hangzhou, China (30.3012 N, 120.0866 E). The specimen was deposited at the Crop Disease Laboratory, Zhejiang Academy of Agricultural Sciences in China, under voucher CDL01018 (Jiaoyu Wang, [email protected]).
Methods
Mycelial growth on the CM medium was collected using the documented method (Xu et al. Citation2011). DNA was extracted using the CTAB method (Talbot et al. Citation1993). Sequencing was performed on an Illumina HiSeq 2000 platform (Illumina, San Diego, CA). Two DNA libraries, PE150 with an insert length of 500 bp (SRR20852133) and PE250 with an insert length of 500 bp (SRR20852132), a stranded RNA-Seq library with an insert length of 300 bp (SRR25783615), were constructed.
Reads were trimmed and assembled using CLC Genomics Workbench 22 (QIAGEN, DK) with default parameters. Contigs with high coverage and longer than 6 kb were filtered and blasted against the nt database (ftp://ftp.ncbi.nlm.nih.gov/blast/db/FASTA/nt.gz) of NCBI with a cutoff e-value of 10−20. Mitogenome hit was checked for a ring-shaped structure. Overlap of the ends was manually fixed by mapping the reads back to the mitogenome in CLC Genomics Workbench 22. Further, the resulting contig was annotated using the MITOS2 web service with RefSeq 63 Fungi dataset and mold genetic code table (Donath et al. Citation2019). The detailed annotation manual curation process using infernal with Rfam (Nawrocki and Eddy Citation2013, Kalvari et al. Citation2021), Prokka (Seemann Citation2014), hmmer with Pfam (Eddy Citation2011, Mistry et al. Citation2021) was provided in Figure S1. The sequences used in the phylogenetic analysis were rotated by MARS with the ‘-a DNA’ option (Ayad and Pissis Citation2017) and aligned using MAFFT version 7.490 (Katoh et al. Citation2002, Katoh and Standley Citation2013). The aligned sequences were trimmed using trimAL 1.4.rev15 with the ‘-automated1’ option (Capella-Gutierrez et al. Citation2009). Nucleotide substitution saturation test was performed using DAMBE 7.3.5 (Xia Citation2018). A maximum likelihood (ML) tree was constructed using RAxML version 8.2.12 (Stamatakis Citation2014) with 1000 rapid bootstraps, GTRGAMMA model was employed.
Results
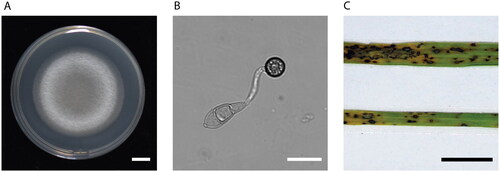
The typical colony morphology, appressorium structure, and blast symptoms on rice leaves of P. oryzae are shown in . The pathogen forms an appressorium, the specialized structure, to penetrate the leaf cuticles.
Figure 1. Colony, appressorium morphology, and blast symptoms. (A) Mycelium growth on a petri dish (Photo taken by Jiaoyu Wang). (B) The three-celled conidia germinate and form an appressorium, the specialized penetration structure (Photo taken by Jiaoyu Wang). (C) Rice leaves with rice blast symptoms (Photo taken by Xiaohong Liu). scale bars are 1 cm, 10 µm, and 1 cm, respectively.
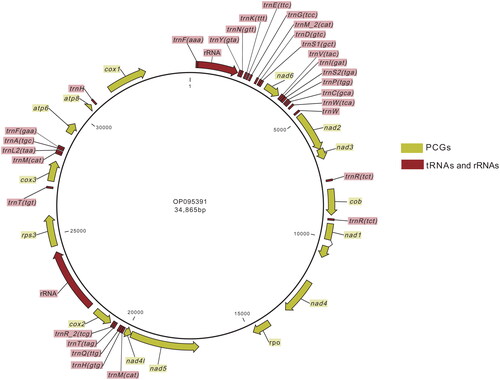
About 71M reads with 13 G bases were used in the assembling. After filtration and identification, the resulting contig has an average coverage of 6555.32. By mapping the reads to the manually curated mitogenome sequence, per-site coverage was determined after deduplication. It varies from 6× to 12635× (Figure S2). Sites with coverage less than 30× were manually checked (Table S1). No errors or heterozygous sites were found in the sequence. The final complete mitogenome of P. oryzae Guy11 (GenBank accession number OP095391) is 34,865 bp in length, with a double-stranded molecular weight of 21.536 MDa (). The overall base composition was 34.9% A, 13.4% C, 15.2% G, and 36.5% T, with a biased GC content of 28.6%. The AT skew is −0.022, and the GC skew is 0.064, indicating a higher Ts than As, and Gs than Cs, respectively. The genome includes 29 tRNAs, 2 rRNAs, and 15 protein-coding genes (PCGs). The lengths of the tRNAs vary from 58 to 85 bp. Small and large rRNA genes are 1588 and 2542 bp, respectively. All of the PCGs use the typical start codon of ATG. cox2, nad1, and rps3 use the stop codon of TAG, while others use TAA. nad1 is confirmed as a split gene with two exons in the manual curation process of annotations (Figure S3).
Figure 2. Genome map of Pyricularia oryzae Guy11 mitochondrion. This map was generated by CLC Genomics Workbench. Features were individually labeled.
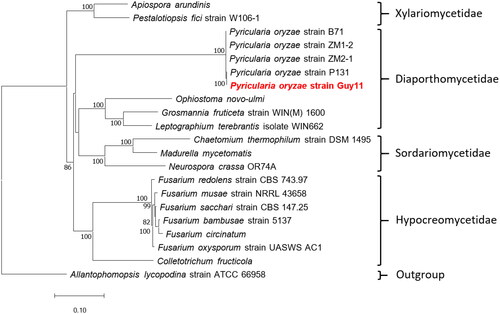
As shown in , based on the complete mitochondrial genome sequence of 21 species, the 20 Sordariomycetes species form a monophyletic group using Allantophomopsis lycopodina from Leotiomycetes as the outgroup. Currently, there are about 761 complete Sordariomycetes species mitochondrial genomes in GenBank, 701 of which are from Hypocreomycetidae. All five available mitogenomes of different P. oryzae strains are involved in the analysis. They are two rice strains, Guy11 (this study) and P131 (Yoshida et al. Citation2016); three wheat strains, B71 (Peng et al. Citation2019), ZM1-2, and ZM2-1. They are also the all available mitogenomes of Magnaporthales species. Besides, Xylariomycetidae and Hypocreomycetidae are separated from other Sordariomycetes species in the analysis.
Figure 3. Phylogenetic tree of maximum likelihood (ML) method based on the mitogenome sequences of Pyricularia oryzae Guy11 and 19 Sordariomycetes species. The tree is rooted with Allantophomopsis lycopodina strain ATCC 66958 (Chen et al. Citation2023). bootstrap support values based on 1,000 replicates are displayed on each node as >70. The following sequences were used: Pyricularia oryzae strain ZM1-2 CM048866 (unpublished), Pyricularia oryzae strain B71 CP060338 (Peng et al. Citation2019), Pyricularia oryzae strain P131 CP114142 (unpublished), Pyricularia oryzae strain Guy11 OP095391 (this study), Pyricularia oryzae strain ZM2-1 CP099704 (unpublished), Allantophomopsis lycopodina strain ATCC 66958 CP103019 (unpublished), chaetomium thermophilum strain DSM 1495 JN007486 (Amlacher et al. Citation2011), madurella mycetomatis JQ015302 (van de Sande Citation2012), Fusarium circinatum JX910419 (Fourie et al. Citation2013), Neurospora crassa OR74A KC683708 (unpublished), Fusarium oxysporum strain UASWS AC1 KR952337 (unpublished), colletotrichum fructicola KX034082 (Liang et al. Citation2017), pestalotiopsis fici strain W106-1 KX870077 (Zhang et al. Citation2017), apiospora arundinis KY775582 (unpublished), ophiostoma novo-ulmi MG020143 (Abboud et al. Citation2018), Fusarium bambusae strain 5137 MH684411 (Wang et al. Citation2018), Fusarium redolens strain CBS 743.97 MT010909 (Yang et al. Citation2020), Fusarium sacchari strain CBS 147.25 MT010910 (Yang et al. Citation2020), Fusarium musae strain NRRL 43658 ON240982 (Degradi et al. Citation2022), leptographium terebrantis isolate WIN662 OP973818 (unpublished), grosmannia fruticeta strain WIN(M) 1600 OQ851465 (unpublished).
Discussion and conclusion
In this study, we got the complete mitochondrion sequence of P. oryzae strain Guy11, the most widely studied strain in this species. There are five complete mitochondrial sequences of different P. oryzae strains currently available in the International Nucleotide Sequence Database Collaboration (INSDC). They form a clade apart from other Diaporthomycetidae and Sordariomycetidae species. Diaporthomycetidae divided from Sordariomycetidae since 2015 based on the morphology and combined analysis of LSU, SSU, TEF, and RPB2 sequence data (Maharachchikumbura et al. Citation2015). This classification is becoming popular in recent studies (Chen et al. Citation2023, Wang et al. Citation2023). Diapothomycetidae is not accepted as a separate subclass apart from Sordariomycetidae in NCBI taxonomy. In this analysis, the resolution between Diaporthomycetidae and Sordariomycetidae is limited due to the need for more data from transitional species. Given the economic importance of P. oryzae in the global agricultural system, the complete mitochondrial sequence of strain Guy11 will be a valuable resource for species identification and population genetics, as well as phylogenetics and comparative genomics in Sordariomycetes and Magnaporthales.
Ethical approval
No ethical issues were involved in this study. The collection and storage of the material were legal and reasonable. Information on the voucher specimen and who identified it were included in the manuscript.
Author contributions
FX conceptualized the study design, performed bioinformatics analysis, and drafted the manuscript. XHL collected and cultured the fungus. JYW analyzed the fungus. XHL and JYW critically revised the manuscript for intellectual content. All authors approved the final manuscript and agreed to be accountable for all aspects of the work.
Supplemental Material
Download MS Word (138.5 KB)Supplemental Material
Download MS Word (635.2 KB)Supplemental Material
Download MS Word (5.6 MB)Supplemental Material
Download MS Word (16.2 KB)Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The data supporting this study’s findings are openly available in the NCBI GenBank (https://www.ncbi.nlm.nih.gov/genbank/) with accession number OP095391. The associated BioProject, BioSample, and SRA numbers are PRJNA866295, SAMN30160189/SAMN37189429, and SRR20852133/SRR20852132/SRR25783615.
Additional information
Funding
References
- Abboud TG, Zubaer A, Wai A, Hausner G. 2018. The complete mitochondrial genome of the Dutch elm disease fungus Ophiostoma novo-ulmi subsp. novo-ulmi. Can J Microbiol. 64(5):339–348. doi: 10.1139/cjm-2017-0605.
- Amlacher S, Sarges P, Flemming D, van Noort V, Kunze R, Devos DP, Arumugam M, Bork P, Hurt E. 2011. Insight into structure and assembly of the nuclear pore complex by utilizing the genome of a eukaryotic thermophile. Cell. 146(2):277–289. doi: 10.1016/j.cell.2011.06.039.
- Ayad LA, Pissis SP. 2017. MARS: improving multiple circular sequence alignment using refined sequences. BMC Genomics. 18(1):86. doi: 10.1186/s12864-016-3477-5.
- Capella-Gutierrez S, Silla-Martinez JM, Gabaldon T. 2009. trimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics. 25(15):1972–1973. doi: 10.1093/bioinformatics/btp348.
- Chao CCT, Ellingboe AH. 1991. Selection for mating competence in Magnaporthe grisea pathogenic to rice. Can J Bot. 69(10):2130–2134. doi: 10.1139/b91-267.
- Chen Y, Su P, Hyde K, Maharachchikumbura S. 2023. Phylogenomics and diversification of Sordariomycetes. Mycosphere. 14(1):414–451. doi: 10.5943/mycosphere/14/1/5.
- Dean RA, Talbot NJ, Ebbole DJ, Farman ML, Mitchell TK, Orbach MJ, Thon M, Kulkarni R, Xu JR, Pan H, et al. 2005. The genome sequence of the rice blast fungus Magnaporthe grisea. Nature. 434(7036):980–986. doi: 10.1038/nature03449.
- Degradi L, Tava V, Prigitano A, Esposto MC, Tortorano AM, Saracchi M, Kunova A, Cortesi P, Pasquali M. 2022. Exploring mitogenomes diversity of Fusarium musae from banana fruits and human patients. Microorganisms. 10(6):1115. doi: 10.3390/microorganisms10061115.
- Donath A, Juhling F, Al-Arab M, Bernhart SH, Reinhardt F, Stadler PF, Middendorf M, Bernt M. 2019. Improved annotation of protein-coding genes boundaries in metazoan mitochondrial genomes. Nucleic Acids Res. 47(20):10543–10552. doi: 10.1093/nar/gkz833.
- Eddy SR. 2011. Accelerated profile HMM searches. PLOS Comput Biol. 7(10):e1002195. doi: 10.1371/journal.pcbi.1002195.
- Fourie G, van der Merwe NA, Wingfield BD, Bogale M, Tudzynski B, Wingfield MJ, Steenkamp ET. 2013. Evidence for inter-specific recombination among the mitochondrial genomes of Fusarium species in the Gibberella fujikuroi complex. BMC Genomics. 14(1):605. doi: 10.1186/1471-2164-14-605.
- Kalvari I, Nawrocki EP, Ontiveros-Palacios N, Argasinska J, Lamkiewicz K, Marz M, Griffiths-Jones S, Toffano-Nioche C, Gautheret D, Weinberg Z, et al. 2021. Rfam 14: expanded coverage of metagenomic, viral and microRNA families. Nucleic Acids Res. 49(D1):D192–D200. doi: 10.1093/nar/gkaa1047.
- Katoh K, Misawa K, Kuma K, Miyata T. 2002. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 30(14):3059–3066. doi: 10.1093/nar/gkf436.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780. doi: 10.1093/molbev/mst010.
- Klaubauf S, Tharreau D, Fournier E, Groenewald JZ, Crous PW, de Vries RP, Lebrun MH. 2014. Resolving the polyphyletic nature of Pyricularia (Pyriculariaceae). Stud Mycol. 79(1):85–120. doi: 10.1016/j.simyco.2014.09.004.
- Leung H, Borromeo ES, Bernardo MA, Notteghem JL. 1988. Genetic analysis of virulence in the rice blast fungus Magnaporthe grisea. Phytopathology. 78(9):1227–1233. doi: 10.1094/Phyto-78-1227.
- Liang X, Tian X, Liu W, Wei T, Wang W, Dong Q, Wang B, Meng Y, Zhang R, Gleason ML, et al. 2017. Comparative analysis of the mitochondrial genomes of Colletotrichum gloeosporioides sensu lato: insights into the evolution of a fungal species complex interacting with diverse plants. BMC Genomics. 18(1):171. doi: 10.1186/s12864-016-3480-x.
- Maharachchikumbura SSN, Hyde KD, Jones EBG, McKenzie EHC, Huang S-K, Abdel-Wahab MA, Daranagama DA, Dayarathne M, D’souza MJ, Goonasekara ID, et al. 2015. Towards a natural classification and backbone tree for Sordariomycetes. Fungal Diversity. 72(1):199–301. doi: 10.1007/s13225-015-0331-z.
- Mistry J, Chuguransky S, Williams L, Qureshi M, Salazar GA, Sonnhammer ELL, Tosatto SCE, Paladin L, Raj S, Richardson LJ, et al. 2021. Pfam: the protein families database in 2021. Nucleic Acids Res. 49(D1):D412–D419. doi: 10.1093/nar/gkaa913.
- Nalley L, Tsiboe F, Durand-Morat A, Shew A, Thoma G. 2016. Economic and environmental impact of rice blast pathogen (Magnaporthe oryzae) alleviation in the United States. PLOS One. 11(12):e0167295. doi: 10.1371/journal.pone.0167295.
- Nawrocki EP, Eddy SR. 2013. Infernal 1.1: 100-fold faster RNA homology searches. Bioinformatics. 29(22):2933–2935. doi: 10.1093/bioinformatics/btt509.
- Peng Z, Oliveira-Garcia E, Lin G, Hu Y, Dalby M, Migeon P, Tang H, Farman M, Cook D, White FF, et al. 2019. Effector gene reshuffling involves dispensable mini-chromosomes in the wheat blast fungus. PLOS Genet. 15(9):e1008272. doi: 10.1371/journal.pgen.1008272.
- Pordel A, Ravel S, Charriat F, Gladieux P, Cros-Arteil S, Milazzo J, Adreit H, Javan-Nikkhah M, Mirzadi-Gohari A, Moumeni A, et al. 2021. Tracing the origin and evolutionary history of Pyricularia oryzae infecting maize and barnyard grass. Phytopathology. 111(1):128–136. doi: 10.1094/PHYTO-09-20-0423-R.
- Seemann T. 2014. Prokka: rapid prokaryotic genome annotation. Bioinformatics. 30(14):2068–2069. doi: 10.1093/bioinformatics/btu153.
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30(9):1312–1313. doi: 10.1093/bioinformatics/btu033.
- Talbot NJ, Salch YP, Ma M, Hamer JE. 1993. Karyotypic variation within clonal lineages of the rice blast fungus, Magnaporthe grisea. Appl Environ Microbiol. 59(2):585–593. doi: 10.1128/aem.59.2.585-593.1993.
- van de Sande WW. 2012. Phylogenetic analysis of the complete mitochondrial genome of Madurella mycetomatis confirms its taxonomic position within the order Sordariales. PLOS One. 7(6):e38654. doi: 10.1371/journal.pone.0038654.
- Wang Z, Kim W, Wang YW, Yakubovich E, Dong C, Trail F, Townsend JP, Yarden O. 2023. The Sordariomycetes: an expanding resource with Big Data for mining in evolutionary genomics and transcriptomics. Front Fungal Biol. 4:1214537. doi: 10.3389/ffunb.2023.1214537.
- Wang XC, Zeng ZQ, Zhuang WY. 2018. The complete mitochondrial genome of the bambusicolous fungus Fusarium bambusae (Nectriaceae, Ascomycota). Mitochondrial DNA B Resour. 3(2):1147–1148. doi: 10.1080/23802359.2018.1522979.
- Xia X. 2018. DAMBE7: new and improved tools for data analysis in molecular biology and evolution. Mol Biol Evol. 35(6):1550–1552. doi: 10.1093/molbev/msy073.
- Xu F, Liu XH, Zhuang FL, Zhu J, Lin FC. 2011. Analyzing autophagy in Magnaporthe oryzae. Autophagy. 7(5):525–530. doi: 10.4161/auto.7.5.15020.
- Yang M, Zhang H, van der Lee TAJ, Waalwijk C, van Diepeningen AD, Feng J, Brankovics B, Chen W. 2020. Population genomic analysis reveals a highly conserved mitochondrial genome in Fusarium asiaticum. Front Microbiol. 11:839. doi: 10.3389/fmicb.2020.00839.
- Yoshida K, Saunders DG, Mitsuoka C, Natsume S, Kosugi S, Saitoh H, Inoue Y, Chuma I, Tosa Y, Cano LM, et al. 2016. Host specialization of the blast fungus Magnaporthe oryzae is associated with dynamic gain and loss of genes linked to transposable elements. BMC Genomics. 17(1):370. doi: 10.1186/s12864-016-2690-6.
- Zhang S, Wang XN, Zhang XL, Liu XZ, Zhang YJ. 2017. Complete mitochondrial genome of the endophytic fungus Pestalotiopsis fici: features and evolution. Appl Microbiol Biotechnol. 101(4):1593–1604. doi: 10.1007/s00253-017-8112-0.
- Zhang H, Zheng X, Zhang Z. 2016. The Magnaporthe grisea species complex and plant pathogenesis. Mol Plant Pathol. 17(6):796–804. doi: 10.1111/mpp.12342.
