Abstract
Scutellaria barbata D. Don 1825 is an important medicinal plant distributed in wetlands about 2000 m above sea level and used to treat various diseases. The complete chloroplast genome of S. barbata is 152,050 bp with four subregions consisting of a large single-copy region (84,053 bp), a small single-copy region (17,517 bp), and a pair of inverted repeats (25,240 bp). In the chloroplast genome of S. barbata, 131 genes were detected, comprising 87 protein-encoding genes, eight ribosomal RNA (rRNA) genes, and 36 transfer RNA (tRNA) genes. Phylogenetic analysis based on the complete chloroplast genome and protein-coding DNA sequences of 27 related taxa of the genus (out group included Holmskioldia sanguinea and Tinnea aethiopica) indicates that S. barbata was made a clade with S. orthocalyx, and S. meehanioides was a sister to them. The first chloroplast genome of S. barbata was reported in this work, serving as a potential reference for important medicinal plants within the Scutellaria genus.
Introduction
Scutellaria barbata D. Don is a perennial herb that often grows on paddy fields, streams, or wet grasslands, with an altitude below 2000 m. The genus Scutellaria was classified into five distinct sections, namely Scutellaria (Rech.) Paton, Anaspis (Rech.) Paton, Salazaria (Torrey) Paton, Perilomia (Kunth) Epling emend. Paton, and Salviifoliae (Boiss.) Edmondson (Ranjbar and Mahmoudi Citation2013). There are more than 300 species of plants of Scutellaria Linn. of Labiatae in angiosperms (Bruno et al. Citation2002), distributed worldwide. The Irano-Turanian region, specifically Central Asia and Afghanistan, represents the primary hub of maximal diversity for the genus Scutellaria. Additionally, the Eastern Mediterranean and the Andes serve as secondary centers for its speciation. In China, Scutellaria plants have been used for clearing away heat and toxic materials, inducing diuresis, and treating hepatitis, appendicitis, and traumatic injury, which has been used since 2000 years ago (Shen et al. Citation2021). S. barbata is a traditional Chinese medicine, and its chloroplast genome has not been published, which leads to its systematic genetic location being unclear. For this reason, we constructed a high-quality assembled chloroplast to enhance the molecular investigation of germplasm, genetic diversity, and phylogenetic relationships.
Materials and methods
Fresh leaf samples of S. barbata D. Don were collected from Mountain Jinyun, Chongqing (Geospatial coordinates: N29.83, E106.39). The original plant was identified as S. barbata by Professor Quan Zhang and the pictures of the original plants were taken by ourselves (). Total DNA was isolated from fresh leaves of the species using a Plant Genomic DNA kit (Tiangen Biotech, Beijing, China). The genome sequence was performed on the Hiseq 2500 platform (Illumina, San Diego, CA). A specimen was deposited at the Guizhou Tobacco Company Anshun Tobacco Company in Guizhou (Prof. Quan Zhang, E-mail: [email protected]) under the voucher number 202210122.
Figure 1. Photographs of Scutellaria barbata D. Don. (These photographs were taken by Prof. Quan Zhang). The foliage of S. barbata exhibits triangular, ovate, or ovate-lanceolate shapes, characterized by a sharp apex and a broad wedge-shaped or nearly truncated base. The raceme of this species is inconspicuous and positioned terminally. The lower bracts are elliptic or narrowly elliptic, while the bracteoles take the form of needle-shaped structures. Furthermore, the corollas of Scutellaria barbata display a vivid purple-blue coloration. (A) Plant panorama of S. barbata and (B) the flowers of S. barbata.

The raw data, consisting of 5.80 GB of raw data, comprised a total of 19,200,779 reads. These reads were subsequently utilized in the assembly of a chloroplast genome, employing the NOVOPlasty (version 2.7.2) (Dierckxsens et al. Citation2017). Annotation was performed using CPGAVAS2 (Shi et al. Citation2019). The genome sequence was confirmed by aligning all raw reads against the assembled genome using BWA v0.7.17 and SAMtools v1.9 (WGS500 Consortium 2014) in the environment of Genome Information System (GeIS; https://geis.infoboss.co.kr/).
To explore the phylogenomic relationship of Scutellaria, 26 complete chloroplast genome sequences were downloaded, including two outgroup plants. The common genes of 27 chloroplast genomes (including S. barbata, and out group were H. sanguinea and T. aethiopica) were extracted and concatenated with Phylosuite (Zhang et al. Citation2020). These sequences were aligned using MAFFT (v7.450) (Rozewicki et al. Citation2019), and a phylogenetic tree was constructed using the alignment and ML method implemented in IQtree (Trifinopoulos et al. Citation2016).
Results
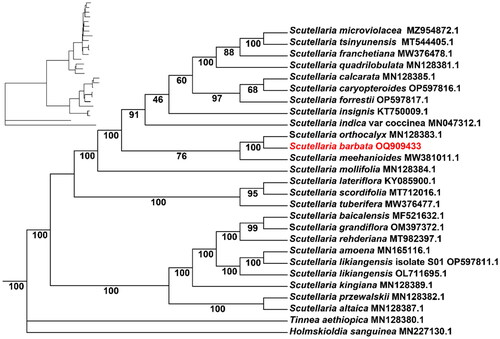
The average and minimum read mapping depths of the assembled genome were 2099× and 540×, respectively (Figure S1). A circular map of the chloroplast genome and a schematic map of the cis- and trans-splicing genes ( and FigureS2) were visualized by CPGView (Liu et al. Citation2023). The total length of the chloroplast genome of S. barbata (GenBank accession number: NC_059814.1) is 152,050 bp with 38.37% GC content (). The chloroplast genome of S. barbata is a typical quadratic structure containing a pair of inverted repeats (IRs) of 25,240 bp separated by a large single-copy (LSC) region of 84,053 bp and a small single-copy (SSC) region of 17,517 bp. The chloroplast genome included 131 genes, of which 112 are unique genes, including 87 protein-coding genes, eight ribosomal RNA (rRNA), and 36 transfer RNA (tRNA) genes. Nineteen protein-coding genes had one intron, and four had two introns. The phylogenetic analysis revealed that S. barbata was taxonomically classified within the genus Scutellaria. Furthermore, it was observed that S. barbata formed a monophyletic clade together with S. orthocalyx, while S. meehanioides was found to be their sister taxon (). The chloroplast genome sequence of S. barbata is a valuable resource for genome evolution and taxonomy research of the genus Scutellaria.
Figure 2. The circle map of chloroplast genome map of S. barbata. Distinctive colored boxes encircling the outer circle depict genes, with clockwise and counter-clockwise transcribed genes represented inside and outside the circle, respectively. The inner circle features a gray region indicating the GC content, while the quadripartite structure (LSC, SSC, IRA, and IRB) is illustrated on the inner circle accordingly.
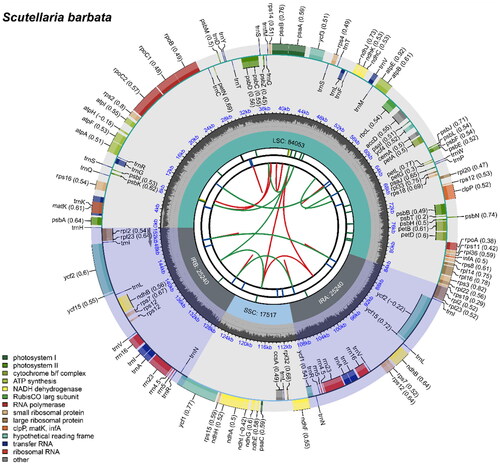
Figure 3. The maximum-likelihood phylogenetic tree of 25 Scutellaria species was constructed based on the CDS sequences extracted by IQ-TREE, with Tinnea aethiopica and Holmskioldia sanguinea added as outgroup. The phylogenetic tree was constructed using the maximum-likelihood method (ML) and bootstrap was performed 1000 times. The number on each branch indicates the boot support value. The following sequences were used: S. microviolacea MZ954872.1 (Wang et al. Citation2022), S. tsinyunensis MT544405.1 (Shan et al. Citation2021), S. franchetiana MW376478.1, S. calcarata MN128385.1 (Zhao et al. Citation2020), S. quadrilobulata MN128381.1 (Zhao et al. Citation2020), S. caryopteroides OP597816.1, S. forrestii OP597817.1, S. insignis KT750009.1, S. indica var coccinea MN047312.1 (Lee and Kim Citation2019), S. orthocalyx MN128383.1 (Zhao et al. Citation2020), S. meehanioides MW381011.1 (Zhang et al. Citation2021), S. mollifolia MN128384.1 (Zhao et al. Citation2020), S. lateriflora KY085900.1, S. scordifolia MT712016.1, S. tuberifera MW376477.1 (Shan et al. Citation2021), S. baicalensis MF521632.1 (Jiang et al. Citation2017), S. grandiflora OM397372.1, S. rehderiana MT982397.1, S. amoena MN165116.1 (Chen and Zhang Citation2019), S. likiangensis isolate S01 OP597811.1, S. likiangensis OL711695.1, S. kingiana MN128389.1 (Zhao et al. Citation2020), S. przewalskii MN128382.1 (Zhao et al. Citation2020), S. altaica MN128387.1 (Zhao et al. Citation2020), T. aethiopica MN128380.1 (Zhao et al. Citation2020), and H. sanguinea MN227130.1 (Lee and Kim Citation2020).
Discussion and conclusions
The complete chloroplast genome of S. barbata was first sequenced and found to exhibit a total length of 152,050 bp. The genome size and gene content of S. barbata are not significantly different from those of most chloroplast genomes or plastomes in the genus Scutellaria (Chen Citation2019; Shan et al. Citation2021). Our research results could be used for authenticating S. barbata and analyzing the genetic diversity and phylogenetic relationships in the genus Scutellaria. Although the species of the genus Scutellaria are very rich, the Scutellaria plant sequence is still relatively few in NCBI (latest date: 25 May 2023). Therefore, to further study the evolutionary history of S. barbata, a more complete Scutellaria species chloroplast sequence is required. The unveiling of the chloroplast genome sequence of S. barbata will provide meaningful information for the phylogeny and plant molecular identification of Scutellaria species.
Author contributions
Shouhui Pan: drafting the work and revising it critically for important intellectual content. Xiquan Li and Li Zhang: analyzed and interpretated the data for the work. Quan Zhang: final check, revision, and approval of the version to be published. Authors agree to be accountable for all aspects of the work.
Ethical approval
The authors declare no ethical or legal violations when obtaining the study materials and performing the research. The species used in this study is not listed on the IUCN Red List, and the sample was legally collected by guidelines stipulated in national and international regulations. The materials were collected in a location not designated as a protected area in China.
Supplemental Material
Download MS Word (198.8 KB)Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The genome sequence data supporting the findings of this study can be publicly obtained at NCBI GenBank in https://www.ncbi.nlm.nih.gov with the accession number OQ909433. Associated BioProject, SRA, and Bio-Sample numbers are PRJNA947595, SRR23952789, and SAMN33861697.
Additional information
Funding
References
- Bruno M, Piozzi F, Maggio AM, Simmonds MS. 2002. Antifeedant activity of neoclerodane diterpenoids from two Sicilian species of Scutellaria. Biochem Syst Ecol. 30(8):793–799. doi: 10.1016/S0305-1978(01)00143-0.
- Chen Q, Zhang D. 2019. Characterization of the complete chloroplast genome of Scutellaria amoena CH Wright (Lamiaceae), a medicinal plant in southwest China. Mitochondrial DNA B Resour. 4(2):3057–3059. doi: 10.1080/23802359.2019.1666677.
- Chen S. 2019. Genetic and phylogenetic analysis of the complete genome for the herbal medicine plant of Scutellaria baicalensis from China. Mitochondrial DNA B Resour. 4(1):1683–1685. doi: 10.1080/23802359.2019.1605859.
- Dierckxsens N, Mardulyn P, Smits G. 2017. NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 45(4):e18. doi: 10.1093/nar/gkw955.
- Jiang D, Zhao Z, Zhang T, Zhong W, Liu C, Yuan Q, Huang L. 2017. The chloroplast genome sequence of Scutellaria baicalensis provides insight into intraspecific and interspecific chloroplast genome diversity in Scutellaria. Genes. 8(9):227. doi: 10.3390/genes8090227.
- Lee Y, Kim S. 2019. The complete chloroplast genome of Scutellaria indica var. coccinea (Lamiaceae), an endemic taxon in Korea. Mitochondrial DNA B Resour. 4(2):2539–2540. doi: 10.1080/23802359.2019.1640649.
- Lee Y, Kim S. 2020. The complete chloroplast genome sequence Holmskioldia sanguinea retz., an ornamental plant of Lamiaceae. Mitochondrial DNA B Resour. 5(1):895–896. doi: 10.1080/23802359.2020.1717392.
- Liu S, Ni Y, Li J, Zhang X, Yang H, Chen H, Liu C. 2023. CPGView: a package for visualizing detailed chloroplast genome structures. Mol Ecol Resour. 23(3):694–704. doi: 10.1111/1755-0998.13729.
- Ranjbar M, Mahmoudi C. 2013. Chromosome numbers and biogeography of the genus Scutellaria L. (Lamiaceae). Caryologia. 66(3):205–214. doi: 10.1080/00087114.2013.821840.
- Rimmer A, Phan H, Mathieson I, Iqbal Z, Twigg SRF, Wilkie AOM, McVean G, Lunter G, WGS500 Consortium. 2014. Integrating mapping-, assembly- and haplotype-based approaches for calling variants in clinical sequencing applications. Nat Genet. 46(8):912–918. doi: 10.1038/ng.3036.
- Rozewicki J, Li S, Amada KM, Standley DM, Katoh K. 2019. MAFFT-DASH: integrated protein sequence and structural alignment. Nucleic Acids Res. 47(W1):W5–W10. doi: 10.1093/nar/gkz342.
- Shan Y, Pei X, Yong S, Li J, Qin Q, Zeng S, Yu J. 2021. Analysis of the complete chloroplast genomes of Scutellaria tsinyunensis and Scutellaria tuberifera (Lamiaceae). Mitochondrial DNA B Resour. 6(9):2672–2680. doi: 10.1080/23802359.2021.1920491.
- Shen J, Li P, Liu S, Liu Q, Li Y, Sun Y, He C, Xiao P. 2021. Traditional uses, ten-years research progress on phytochemistry and pharmacology, and clinical studies of the genus Scutellaria. J Ethnopharmacol. 265:113198. doi: 10.1016/j.jep.2020.113198.
- Shi L, Chen H, Jiang M, Wang L, Wu X, Huang L, Liu C. 2019. CPGAVAS2, an integrated plastome sequence annotator and analyzer. Nucleic Acids Res. 47(W1):W65–W73. doi: 10.1093/nar/gkz345.
- Trifinopoulos J, Nguyen L-T, von Haeseler A, Minh BQ. 2016. W-IQ-TREE: a fast online phylogenetic tool for maximum likelihood analysis. Nucleic Acids Res. 44(W1):W232–W235.
- Wang Y-C, Zhang Z-R, Yan L-J. 2022. Characterization of the complete plastid genome of Scutellaria microviolacea (Lamiaceae), a species endemic to Yunnan Province of China. Mitochondrial DNA B Resour. 7(5):758–760. doi: 10.1080/23802359.2022.2069521.
- Zhang C, Xia P, Wu R, Mans D. 2021. The complete chloroplast genome of Scutellaria meehanioides (Lamiaceae) from Shaanxi Province, China. Mitochondrial DNA B Resour. 6(6):1685–1686. doi: 10.1080/23802359.2021.1927877.
- Zhang D, Gao F, Jakovlić I, Zou H, Zhang J, Li WX, Wang GT. 2020. PhyloSuite: an integrated and scalable desktop platform for streamlined molecular sequence data management and evolutionary phylogenetics studies. Mol Ecol Resour. 20(1):348–355. doi: 10.1111/1755-0998.13096.
- Zhao F, Li B, Drew BT, Chen Y-P, Wang Q, Yu W-B, Liu E-D, Salmaki Y, Peng H, Xiang C-L, et al. 2020. Leveraging plastomes for comparative analysis and phylogenomic inference within Scutellarioideae (Lamiaceae). PLOS One. 15(5):e0232602. doi: 10.1371/journal.pone.0232602.
