Abstract
Glyptothorax pallozonus Lin, 1934 is a small benthic fish belonging to the Sisoridae family that is distributed in the Dongjiang and Rongjiang Rivers of China. In the present study, we sequenced and characterized the complete mitochondrial genome of G. pallozonus for the first time. The complete mitogenome of G. pallozonus is 16,542 bp in length and includes 13 protein-coding genes (PCGs), 22 transfer RNAs (tRNAs), two ribosomal RNA (rRNAs), and a control region (CR). The mitogenome architecture was identical to that of other teleosts. Maximum likelihood (ML) phylogenetic analysis strongly supported the monophyly of Glyptothorax, which contains two clades. These results advance our understanding of the molecular phylogeny of the genus Glyptothorax.
Introduction
Glyptothorax is the most species-rich and widely distributed genus in the Sisoridae family and currently comprises 101 recognized species (Bánki et al. Citation2023). Glyptothorax pallozonus is a small benthic fish () that is endemic to the eastern region of Guangdong Province, China, and is characteristically found in the Dongjiang and Rongjiang Rivers (Zheng Citation1991). The species, like other sisorids, live in fast-moving streams, where they have adapted to using an adhesive apparatus on their underside to attach themselves to rocks and prevent themselves from being washed away (Ng and Rachmatika Citation2005). Over the past three decades, G. pallozonus has experienced a decline in population size due to environmental degradation and habitat changes. To date, this species is only occasionally caught in the wild; therefore, studies of this species are limited. Previous studies have mainly focused on the morphological, distributional, and systematic characteristics of G. pallozonus (Zheng Citation1991; Chu and Mo Citation1999; Jiang et al. Citation2011), and genetic information on this species is scarce. In the present study, the complete mitochondrial genome and the phylogenetic relationships of G. pallozonus were determined for the first time.
Materials and methods
A live specimen of G. pallozonus was collected from Fengshun, Guangdong Province, China (23°44′57ʺN, 116°9′26ʺE). The specimen was morphologically identified as described by Zheng (Citation1991) and Chu and Mo (Citation1999). In particular, it could be distinguished from its congeners by the presence of a bright white triangular patch on the base of the dorsal fin (). The specimen was deposited at the ichthyological museum of Freshwater Fisheries Research Institute of Jiangsu Province, China (Dr Liqiang Zhong, e-mail: [email protected]) under the voucher number JSFFRI-18003.
Methods
Genomic DNA was extracted using the Ezup Column Animal Genomic DNA Kit (Sangon, China) following the manufacturer’s protocol. The mitogenome was amplified using 30 sets of fish universal primers (Miya and Nishida Citation1999), and gaps were filled with self-designed primers based on the mitogenomes of other sisorids (Supplementary material, Table S1). The PCR products were sequenced by Sanger sequencing using the same primers.
Raw sequences were manually checked and assembled. The assembled sequences were annotated and visualized using MitoAnnotator (Zhu et al. Citation2023). Based on the concatenated sequences of 13 protein-coding genes (PCGs) from 23 fish (), a maximum likelihood (ML) phylogenetic tree was constructed with 1000 bootstrap assemblies and the GTR + G + I model using MEGA 11 (Tamura et al. Citation2021).
Table 1. Species and GenBank accession number of mitogenomes used in this study.
Results
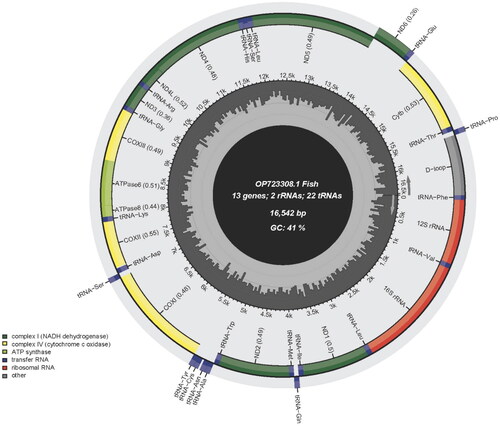
The complete mitochondrial genome of G. pallozonus (GenBank accession number: OP723308) is 16,542 bp long and includes 13 PCGs, two ribosomal RNAs (rRNAs), 22 transfer RNAs (tRNAs), and one control region (CR) (). The mitogenome architecture and gene-coding strands are similar to those of the teleost consensus (Ma et al. Citation2015). We observed a small AT-rich characteristic (59.2%), with a nucleotide composition of 27.1% T, 25.9% C, 32.1% A, and 14.9% G. The ND6 gene and eight tRNAs (tRNAAla, tRNAAsn, tRNACys, tRNAGln, tRNAGlu, tRNAPro, tRNASer, and tRNATyr) were encoded on the light strand, whereas the other components were located on the heavy strand. Twelve PCGs started with ATG, while the COI gene began with GTG. Six genes ended with the complete stop codon TAA (ND1, COI, ATPase8, ND4L, ND5, and ND6), whereas the remaining genes had incomplete stop codons TA (ATPase 6) or T (ND2, COII, COIII, ND3, ND4, and Cytb). The origin of light-strand replication was 32 bp in length and located in the WANCY cluster between tRNAAsn and tRNACys. The CR was 890 bp in length and contained a termination-associated sequence (TAS), three central conserved sequence blocks (CSB-D, CSB-E, and CSB-F), and three conserved sequence blocks (CSB-1, CSB-2, and CSB-3).
Figure 2. Gene map of the mitochondrial genome of Glyptothorax pallozonus (GenBank accession number: OP723308), with 13 protein coding genes, 22 tRNAs, 2 rRNAs, and a control region. Genes encoded on light strand and heavy strand were shown on the inner and outer sides of the ring, respectively.
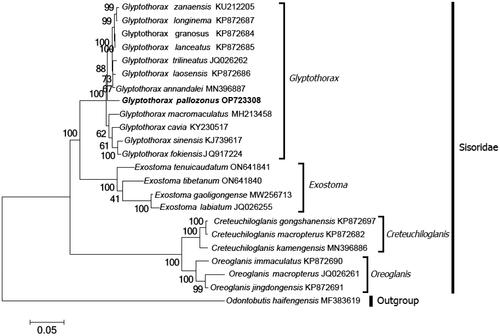
The ML phylogenetic tree indicated the monophyly of the genus Glyptothorax with high support scores (). Two clades were identified within the genus Glyptothorax; G. pallozonus is grouped with G. zanaensis, G. longinema, G. granosus, G. lanceatus, G. trilineatus, G. laosensis, and G. annandalei, and eventually forms a clade. The remaining four species, G. macromaculatus, G. cavia, G. sinensis, and G. fokiensis, form a separate clade.
Figure 3. Maximum-likelihood (ML) phylogenetic tree was reconstructed based on the concatenated 13 protein-coding genes of Glyptothorax pallozonus and other 22 fishes. Accession numbers were indicated after the species name. Numbers at the nodes indicated bootstrap support values from 1000 replicates.
Discussion and conclusion
The structure of the complete mitochondrial genome of G. pallozonus is similar to that of other teleosts (Miya et al. Citation2003). In particular, the mitogenome structures of the genus Glyptothorax are highly conserved, suggesting that the mitochondrial genome is a perfect tool for population genetics and phylogenetic studies of these fishes. The ML phylogenetic tree strongly supported the monophyly of Glyptothorax, which was consistent with the results of a previous study based on the single gene sequences of RAG2, COI, and Cyt b (Jiang et al. Citation2011). Previous phylogenetic studies have included only four to seven species in the genus Glyptothorax and thus, have generated only a limited phylogeny of these fishes. Based on phylogenetic analysis, these fishes can be divided into two main clades: G. zanaensis and G. trilineatus, and G. macromaculatus, G. sinensis, and G. fokiensis (Huang et al. Citation2017; Li Citation2017; Lv et al. Citation2018). The new mitogenome discovered in this study supports the placement of G. pallozonus in the Glyptothorax genus. To the best of our knowledge, this is the first report on the evolutionary position of this species. At the same time, in the present study, the phylogenies of all 12 available fishes of Glyptothorax with complete mitogenomes were reconstructed, which would be useful in understanding the relationships within the Glyptothorax genus. However, the complete mitochondrial genome of most fishes in Glyptothorax (approximately 89 species) is still unavailable, and to elucidate the phylogenetic relationships of the Glyptothorax genus, extensive sampling and additional molecular information is necessary.
Author contributions
QFG and ZLQ conceived this study; QFG and YS conducted the experiments, LAP and WQ analyzed the data; QFG wrote the drafting of the paper; QFG and ZLQ revised it critically. All authors contributed to the article and approved the submitted version.
Ethical approval
The species used in this study is not protected under CITES or wildlife laws in China, and its status is data deficient by the IUCN. Therefore, no specific permission or license is required for research sampling according to Regulations of the People’s Republic of China. The live samples were obtained in accordance with the guidelines of the animal care and Ethical Committee of Freshwater Fisheries Research Institute of Jiangsu Province and Animal Experiments of Jiangsu Agri-animal Husbandry Vocational College.
Supplemental Material
Download MS Word (21.4 KB)Acknowledgments
We are grateful to Shujie Liu, Binbin Zhan and Huiwen Xiao for help in field assistance and the species reference image.
Data availability statement
The mitochondrial genome sequence is available on GenBank of NCBI at www.ncbi.nlm.nih.gov with the accession number of OP723308.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Bánki O, Roskov Y, Döring M, Ower G, Vandepitte L, Hobern D, Remsen D, Schalk P, DeWalt RE, Keping M, et al. 2023. Catalogue of Life Checklist (Version 2023-02-07). Catalogue of Life doi: 10.48580/dfrq.
- Chu XL, Mo TP. 1999. Sisoridae. In: Chu XL, Zheng BS, Dai DY, editors. Fauna sinica osteichthyes siluriformes. Beijing (China): Science Press; p. 114–181.
- Gong Z, Jiang W, Feng H, Liu Y, Zhu T. 2022. Comparative mitogenomics of Two sympatric catfishes of Exostoma (Siluriformes: Sisoridae) from the lower Yarlung tsangpo River and its application for phylogenetic consideration. Genes (Basel). 13(9):1615. doi: 10.3390/genes13091615.
- Gong Z, Lin F, Luo Z, Chen X. 2021. The complete mitochondrial genome of Exostoma gaoligongense (Siluriformes: Sisoridae) and its phylogenetic analysis within glyptosternine catfishes. Mitochondrial DNA B Resour. 6(4):1424–1425. doi: 10.1080/23802359.2021.1912670.
- Huang FJ, Liu MD, Yu LX, Liu SP. 2017. The complete mitochondrial genome of Glyptothorax laosensis (Siluriformes, Sisoridae). Mitochondrial DNA A DNA Mapp Seq Anal. 28(1):60–61. doi: 10.3109/19401736.2015.1110798.
- Jiang WS, Ng HH, Yang JX, Chen XY. 2011. Monophyly and phylogenetic relationships of the catfish genus Glyptothorax (Teleostei: Sisoridae) inferred from nuclear and mitochondrial gene sequences. Mol Phylogenet Evol. 61(2):278–289. doi: 10.1016/j.ympev.2011.06.018.
- Li B. 2017. The complete mitochondrial genome of Glyptothorax cavia (Siluriformes, Sisoridae, Glyptothorax): genome characterization and phylogenetic analysis. Mitochondrial DNA B Resour. 2(1):259–260. doi: 10.1080/23802359.2017.1307701.
- Li B, Tian ZF, Qin Y, Hao M, Zhang JB. 2016. The complete mitochondrial genome of Glyptothorax zainaensis (Siluriformes, Sisoridae, Glyptothorax): genome characterization and phylogenetic analysis. Mitochondrial DNA B Resour. 1(1):56–57. doi: 10.1080/23802359.2015.1137821.
- Li JS, Peng Y, Zhang SF, Liu YF, Zhang K, Chen J, Zhang H, Zhang C, Liu BJ. 2023. The complete mitochondrial genome of Parachiloglanis hodgarti and its phylogenetic position within Sisoridae. J Ocean Limnol. 41(1):267–279. doi: 10.1007/s00343-021-1319-z.
- Lin XW, Li L, Jin HY, Jin X, Ma B. 2021. Complete mitochondrial genome sequence of Glyptothorax annandalei in the Yarlung Zangbo River, Tibet. Mitochondrial DNA B Resour. 6(4):1397–1398. doi: 10.1080/23802359.2021.1911701.
- Lv Y, Li Y, Ruan Z, Bian C, You X, Yang J, Jiang W, Shi Q. 2018. The complete mitochondrial genome of Glyptothorax macromaculatus provides a well-resolved molecular phylogeny of the Chinese sisorid catfishes. Genes (Basel). 9(6):282. doi: 10.3390/genes9060282.
- Ma QZ, Li L, Lin XW, Jin HY, Jin X, Ma B. 2020. Complete mitochondrial genome sequence of Pareuchiloglanis kamengensis in the Yarlung Zangbo River, Tibet. Mitochondrial DNA B Resour. 5(1):368–369. doi: 10.1080/23802359.2019.1703576.
- Ma XH, Kang JL, Chen WT, Zhou CJ, He SP. 2015. Biogeographic history and high-elevation adaptations inferred from the mitochondrial genome of Glyptosternoid fishes (Sisoridae, Siluriformes) from the southeastern Tibetan Plateau. BMC Evol Biol. 15(1):233. doi: 10.1186/s12862-015-0516-9.
- Miya M, Nishida M. 1999. Organization of the mitochondrial genome of a deep-sea fish, Gonostoma gracile (Teleostei: Stomiiformes): first example of transfer RNA gene rearrangements in bony fishes. Mar Biotechnol. 1(5):416–426. doi: 10.1007/PL00011798.
- Miya M, Takeshima H, Endo H, Ishiguro NB, Inoue JG, Mukai T, Satoh TP, Yamaguchi M, Kawaguchi A, Mabuchi K, et al. 2003. Major patterns of higher teleostean phylogenies: a new perspective based on 100 complete mitochondrial DNA sequences. Mol Phylogenet Evol. 26(1):121–138. doi: 10.1016/S1055-7903(02)00332-9.
- Ng HH, Rachmatika I. 2005. Glyptothorax exodon, a new species of rheophilic catfish from Borneo (Teleostei: Sisoridae). Raffles Bull Zool. 53(2):251–255.
- Tamura K, Stecher G, Kumar S. 2021. MEGA11: molecular evolutionary genetics analysis version 11. Mol Biol Evol. 38(7):3022–3027. doi: 10.1093/molbev/msab120.
- Yan SH, Xiong MH, Que YF, Tang HY, Zhu B, Shao K. 2016. The complete mitochondrial genome of Glyptothorax sinense (Siluriformes, Sisoridae). Mitochondrial DNA A DNA Mapp Seq Anal. 27(2):886–887. doi: 10.3109/19401736.2014.919489.
- Zheng CY. 1991. Sisoridae. In: Pan JH, Zhong L, Zheng CY, Wu HL, Liu JH, editors. The freshwater fishes of Guangdong Province. Guangzhou (China): Guangdong Science and Technology Press; p. 319–322.
- Zhong LQ, Wang MH, Li DM, Tang SK, Zhang TQ, Bian WJ, Chen XH. 2018. Complete mitochondrial genome of Odontobutis haifengensis (Perciformes, Odontobutiae): A unique rearrangement of tRNAs and additional noncoding regions identified in the genus Odontobutis. Genomics. 110(6):382–388. doi: 10.1016/j.ygeno.2017.12.008.
- Zhou CJ, Wang DQ, Yu ML, He SP. 2012. The complete mitochondrial genome of Glyptothorax fukiensis fukiensis (Teleostei, Siluriformes: Sisoridae). Mitochondrial DNA A. 23(6):414–416. doi: 10.3109/19401736.2012.710211.
- Zhu T, Sato Y, Sado T, Miya M, Iwasaki W. 2023. MitoFish, MitoAnnotator, and MiFish pipeline: updates in 10 years. Mol Biol Evol. 40(3):msda035. doi: 10.1093/molbev/msad035.

