Abstract
Plants of the genus Plectranthus are used for the treatment of digestive problems, skin diseases, and allergies, with a wide variety of uses. Here, the complete chloroplast genome sequence of Plectranthus hadiensis (Benth. ex E.Mey) Codd. 1788 was assembled and characterized for the first time. The full length of the chloroplast genome is 152,484 bp, consisting of a small single-copy region of 17,686 bp, a large single-copy region of 83,380 bp, and a pair of inverted repeats of 51,418 bp. The overall GC content is 37.73%. The chloroplast genome contains 131 unique genes, including 87 protein-coding genes, 36 transfer RNA genes, and eight ribosomal RNA genes. Phylogenetic tree construction based on the complete chloroplast genome sequences of 25 species (23 Nepetoideae, two Ajugoideae) of the Lamiaceae family showed that P. hadiensis exhibited the closest relationship with Isodon.
Introduction
The genus Plectranthus (Lamiaceae) has a long history of treating allergies, skin disorders, allergies, and digestive issues (Lukhoba et al. Citation2006). Plectranthus hadiensis var. tomentosus, also known as Plectranthus tomentosus (Benth. ex E.Mey.) Codd. 1788, is a semi-succulent subshrub with aromatic leaves. The plant is native to South Africa and Asia, including South Korea (Ji et al. Citation2019). The natural products of P. hadiensis contain phenolics and essential oils, including terpenoids (Abdel‑Mogib et al. Citation2002). Recent studies demonstrated that P. hadiensis possesses a wide range of biological characteristics, including cytotoxic, antioxidant, antimicrobial, and anti-inflammatory activities (Marques et al. Citation2002; Mothana et al. Citation2010; Kong et al. Citation2013; Sun et al. Citation2022). In the present study, the whole chloroplast genome of P. hadiensis was characterized and assembled for the first time to reveal the genetic taxonomy at the molecular level and provide a foundation for the evolutionary analysis of P. hadiensis within the Lamiaceae family.
Materials and methods
Fresh P. hadiensis leaves were collected from the garden of Zhejiang Agriculture and Forestry University (30°15′23.71″N, 119°43′41.76″E), Zhejiang Province, China (). A specimen was deposited at Shanghai Chenshan Herbarium, Shanghai, China, under the voucher number CSH0065563 (Contact: Binjie Ge, Email: [email protected]). The genomic DNA was extracted by using the cetyltrimethyl ammonium bromide (CTAB) method with minor modification (Montero-Pau et al. Citation2018), and then the DNA concentration, quality, and integrity were determined by using a Qubit Fluorometer (Invitrogen, Carlsbad, CA) and a NanoDrop Spectrophotometer (Thermo Scientific, Waltham, MA). The constructed libraries were sequenced using the Illumina NovaSeq platform by following the standard Illumina protocols (Illumina, San Diego, CA). The library was sequenced into a paired-end sequencing mode under PE400 library. FastQC was used to rapidly assess the quality of sequencing data (http://www.bioinformatics.babraham.ac.uk/projects/fastqc). Raw sequencing data were preprocessed using AdapterRemoval (version 2.1.7) to remove adapter sequences (Lindgreen Citation2012). Then, SOAPec (v2.0) software was used to correct all the reads based on the K-mer frequency to obtain high-quality data (Luo et al. Citation2012). The high-quality reads were assembled with Fast-Plast to obtain the chloroplast genome (https://github.com/mrmckain/Fast-Plast).
The chloroplast genome of P. hadiensis was annotated using Geseq (https://chlorobox.mpimp-golm.mpg.de/geseq.html), and the Genome Map was drawn with CPGview (http://www.1kmpg.cn/cpgview). Finally, the complete chloroplast genome sequences and annotations of P. hadiensis were submitted to GenBank under the accession number OP611428.
Results
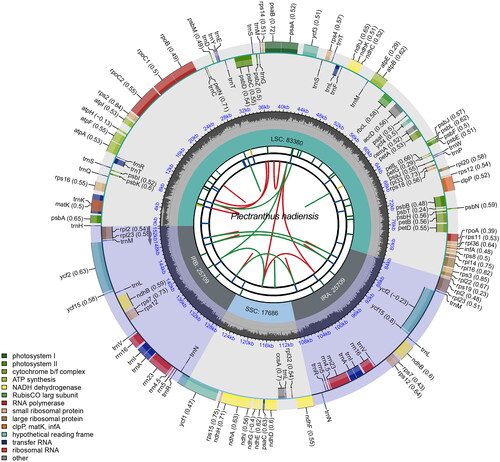
The chloroplast genome of P. hadiensis is a typical circular structure, 152,484 bp in length, consisting of inverted repeat regions (IRa and IRb) of 51,418 bp, a large single-copy (LSC) region of 83,380 bp and a small single-copy (SSC) region of 17,686 bp (). The genome encodes a total of 131 unique genes, including 87 protein-coding genes, 36 transfer RNA genes, and eight ribosomal RNA genes. The total GC content was 37.73%, and the GC contents in LSC, SSC, IRa, and IRb regions were 35.75%, 31.4%, 43.14%, and 43.15%, respectively. As the evidence for correct assembly of the genome, the coverage depth figure is provided in Supplemental Figure S1. The cis-splicing genes and the trans-splicing gene rps12 are shown in Supplemental Figure S2, and the genes containing intron are listed in Supplemental Table S1.
Figure 1. The morphological characteristics of P. hadiensis used in our research. It is a semi-succulent subshrub with aromatic leaves. Its leaves are opposite, light green, broadly ovate with scalloped margins, and densely covered with short hairs. (a) Top view of the P. hadiensis showing the shape of its leaves; (b) front view of the P. hadiensis showing the state of the plant (photos were taken by Jiaojiao Hao in Lin’an District, Hangzhou City, Zhejiang Province, China).

Figure 2. Circular map of the complete chloroplast genome of P. hadiensis generated by CPGview. The map contains six tracks in default. From the center outward, the first track shows the dispersed repeats. The dispersed repeats consist of direct (D) and Palindromic (P) repeats, connected with red and green arcs. The second track shows the long tandem repeats as short blue bars. The third track shows the short tandem repeats or microsatellite sequences as short bars with different colors. The colors, the type of repeat they represent, and the description of the repeat types are as follows: Black: c (complex repeat); green: p1 (repeat unit size = 1); yellow: p2 (repeat unit size = 2); purple: p3 (repeat unit size = 3); blue: p4 (repeat unit size = 4); orange: p5 (repeat unit size = 5); red: p6 (repeat unit size = 6). The small single-copy (SSC), inverted repeat (IRa and IRb), and large single-copy (LSC) regions are shown on the fourth track. The GC content along the genome is plotted on the fifth track. The genes are shown on the sixth track. The optional codon usage bias is displayed in the parenthesis after the gene name. Genes are color-coded by their functional classification. The transcription directions for the inner and outer genes are clockwise and anticlockwise, respectively. The functional classification of the genes is shown in the bottom left corner. Note: The taxonomy name of Plectranthus hadiensis in BioSample and GenBank is Coleus hadiensis (the heterotypic synonym of Plectranthus hadiensis).
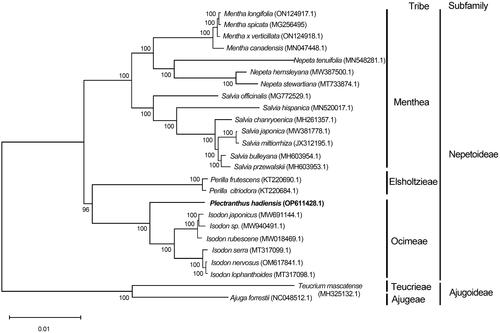
To evaluate the phylogenetic position of P. hadiensis, chloroplast genomes of 25 species (23 Nepetoideae, two Ajugoideae) belonging to the family of Lamiaceae were downloaded from GenBank (Supplemental Table S2). Sequence alignment was performed using Mafft version 7 software (Katoh and Standley Citation2013), and the maximum-likelihood tree was constructed via MEGA11 software under the Tamura-Nei model with 1000 bootstrap replicates (Tamura et al. Citation2011; Kumar et al. Citation2018). The phylogenetic tree revealed that P. hadiensis was clustered in subfamily Nepetoideae belonging to the tribe Ocimeae, and plants in Plectranthus had a close relationship with Isodon ().
Figure 3. Maximum-likelihood phylogenetic tree based on 25 chloroplast genomes of Lamiaceae. Teucrium mascatense and Ajuga forrestii are outgroups. The position of P. hadiensis is marked in bold. Bootstrap values are shown on each node (N = 1000). Scale bar = 0.01. The following sequences were used: Mentha longifolia ON124917.1 (Zubair Filimban et al. Citation2022), Mentha spicata MG256495 (Wang et al. Citation2017), Mentha x verticillata ON124918.1 (Zubair Filimban et al. Citation2022), Mentha canadensis MN047448.1 (Li et al. 2020), Nepeta tenuifolia MN548281.1 (Wang et al. Citation2021), Nepeta hemsleyana MW387500.1 (Bautista et al. Citation2022), Nepeta stewartiana MT733874.1, Salvia officinalis MG772529.1 (Du et al. Citation2022), Salvia hispanica MN520017.1 (Zhao et al. Citation2020), Salvia chanryoenica MH261357.1 (Ha et al. Citation2018), Salvia japonica MW381778.1 (Liang et al. Citation2019), Salvia miltiorrhiza JX312195.1 (Gao et al. Citation2020), Salvia bulleyana MH603954.1 (Liang et al. Citation2019), Salvia przewalskii MH603953.1 (Liang et al. Citation2019), Perilla frutescens KT220690.1 (Lang et al. Citation2020), Perilla citriodora KT220684.1 (Mo et al. Citation2017), Plectranthus hadiensis OP611428.1, Isodon japonicus MW691144.1 (Wang et al. Citation2022), Isodon sp. MW940491.1, Isodon rubescens MW018469.1 (Yue et al. Citation2021), Isodon serra MT317099.1 (Zhang et al. 2023), Isodon nervosus OM617841.1, Isodon lophanthoides MT317098.1 (Zhang, Ma, et al. Citation2020; Zhang, Xia, et al. Citation2020), Teucrium mascatense MH325132.1 (Khan et al. Citation2019), and Ajuga forrestii NC048512.1 (Tao et al. Citation2019). The genome length is shown in Supplementary Table S2.
Discussion and conclusions
As the center of photosynthesis and energy transduction, chloroplasts play a crucial role in the growth and yield of plants (Yu et al. Citation2014). In this study, the complete plastome of P. hadiensis was sequenced, assembled, and annotated for the first time. The genomic structure of P. hadiensis consists of a pair of IRs, an SSC region, and an LSC region similar to that in the majority of other angiosperms (Liu et al. Citation2022). The phylogenetic results indicated that P. hadiensis exhibited the closest relationship with Isodon in the subfamily Nepetoideae. In conclusion, the study provides a molecular basis in revealing the phylogenetic relationships and providing genetic information for the advancement of systematics research among Lamiaceae species, the exploitation of plants, and resource protection.
Author contributions
Zhujun Zhu and Youjian Yu designed the experiment; Jiaojiao Hao and Yanchi Lu analyzed the data and wrote the manuscript; Menghuan Dang, Rui Xia, and Liai Xu were involved in the interpretation of the data. All authors read and approved the final manuscript before submission.
Ethical approval
P. hadiensis is not an endangered or a protected species, and the sample collection site is neither privately owned nor protected for which no specific permissions were required.
Supplemental Material
Download MS Word (370.4 KB)Supplemental Material
Download JPEG Image (778.3 KB)Supplemental Material
Download JPEG Image (1.6 MB)Supplemental Material
Download JPEG Image (1.1 MB)Disclosure statement
The authors reportedly had no conflict of interest.
Data availability statement
The data of this study are openly accessible in GenBank of NCBI at https://www.ncbi.nlm.nih.gov/ under the accession number OP611428. The associated BioProject, SRA, and BioSample numbers are PRJNA875654, SRR21387184, and SAMN30629428, respectively.
Additional information
Funding
References
- Abdel‑Mogib M, Albar H, Batterjee S. 2002. Chemistry of the genus Plectranthus. Molecules. 7(2):271–301. doi: 10.3390/70200271.
- Bautista MAC, Wen X, Ma C, Tu Y, Chen T. 2022. Complete chloroplast genome sequence of the Tibetan catnip Nepeta hemsleyana Oliver ex Prain (Lamiaceae). Mitochondrial DNA B Resour. 7(3):552–553. doi: 10.1080/23802359.2022.2054737.
- Du Q, Yang H, Zeng J, Chen Z, Zhou J, Sun S, Wang B, Liu C. 2022. Comparative genomics and phylogenetic analysis of the chloroplast genomes in three medicinal Salvia species for bioexploration. Int J Mol Sci. 23(20):12080. doi: 10.3390/ijms232012080.
- Gao C, Wu C, Zhang Q, Zhao X, Wu M, Chen R, Zhao Y, Li Z. 2020. Characterization of chloroplast genomes from two Salvia medicinal plants and gene transfer among their mitochondrial and chloroplast genomes. Front Genet. 11:574962. doi: 10.3389/fgene.2020.574962.
- Ha YH, Choi KS, Choi K. 2018. Characterization of complete chloroplast genome of endemic species of Korea Peninsular, Salvia chanryoenica (Lamiaceae). Mitochondrial DNA B Resour. 3(2):992–993. doi: 10.1080/23802359.2018.1495115.
- Ji H-S, Li H, Mo E-J, Kim U-H, Kim Y-H, Park H-Y, Jeong T-S. 2019. Low-density lipoprotein–antioxidant flavonoids and a phenolic ester from Plectranthus hadiensis var. tomentosus. Appl Biol Chem. 62(1):58. doi: 10.1186/s13765-019-0464-y.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780. doi: 10.1093/molbev/mst010.
- Khan A, Asaf S, Khan AL, Khan A, Al-Harrasi A, Al-Sudairy O, AbdulKareem NM, Al-Saady N, Al-Rawahi A. 2019. Complete chloroplast genomes of medicinally important Teucrium species and comparative analyses with related species from Lamiaceae. PeerJ. 7:e7260. doi: 10.7717/peerj.7260.
- Kong W, Lv D, Li H, et al. 2013. Analysis on volatile components in different parts of Plectranthus hadiensis by SPME-GC–MS. Chin J Pharm Anal. 33(2):241–245.
- Kumar S, Stecher G, Li M, Knyaz C, Tamura K. 2018. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. 35(6):1547–1549. doi: 10.1093/molbev/msy096.
- Lang Y, Zhang Z, Jiang J, Cao T, Tian J. 2020. Characterization of the chloroplast genome of Perilla frutescens (Lamiaceae), an herb plant from China. Mitochondrial DNA Part B. 5(2):1550–1551. doi: 10.1080/23802359.2020.1742597.
- Liang C, Wang L, Lei J, Duan B, Ma W, Xiao S, Qi H, Wang Z, Liu Y, Shen X, et al. 2019. A comparative analysis of the chloroplast genomes of four Salvia medicinal plants. Engineering. 5(5):907–915. doi: 10.1016/j.eng.2019.01.017.
- Lindgreen S. 2012. AdapterRemoval: easy cleaning of next-generation sequencing reads. BMC Res Notes. 5(1):337. doi: 10.1186/1756-0500-5-337.
- Liu H, He W, Zhang X, Jiang Z, Li Q, Xia C, Wang H. 2022. Characterization of the complete chloroplast genome of Veronica arvensis and its phylogenomic inference in Plantaginaceae. Mitochondrial DNA B Resour. 7(11):1928–1932. doi: 10.1080/23802359.2022.2139162.
- Lukhoba C, Simmonds M, Paton A. 2006. Plectranthus: a review of ethnobotanical uses. J Ethnopharmacol. 103(1):1–24. doi: 10.1016/j.jep.2005.09.011.
- Luo R, Liu B, Xie Y, Li Z, Huang W, Yuan J, He G, Chen Y, Pan Q, Liu Y, et al. 2012. SOAPdenovo2: an empirically improved memory-efficient short-read de novo assembler. Gigascience. 1(1):18. doi: 10.1186/2047-217X-1-18.
- Marques CG, Pedro M, Simões MFA, Nascimento MSJ, Pinto MMM, Rodríguez B. 2002. Effect of abietane diterpenes from Plectranthus grandidentatus on the growth of human cancer cell lines. Planta Med. 68(9):839–840. doi: 10.1055/s-2002-34407.
- Mo J-S, Kim K, Lee MH, Lee J-H, Yoon U-H, Kim T-H. 2017. The complete chloroplast genome sequence of Perilla citriodora (Makino) Nakai. Mitochondrial DNA A DNA Mapp Seq Anal. 28(1):131–132. doi: 10.3109/19401736.2015.1111356.
- Montero-Pau J, Blanca J, Bombarely A, Ziarsolo P, Esteras C, Martí-Gómez C, Ferriol M, Gómez P, Jamilena M, Mueller L, et al. 2018. De novo assembly of the zucchini genome reveals a whole-genome duplication associated with the origin of the Cucurbita genus. Plant Biotechnol J. 16(6):1161–1171. doi: 10.1111/pbi.12860.
- Mothana RAA, Abdo SAA, Hasson S, Althawab FMN, Alaghbari SAZ, Lindequist U. 2010. Antimicrobial, antioxidant and cytotoxic activities and phytochemical screening of some Yemeni medicinal plants. Evid Based Complement Alternat Med. 7(3):323–330. doi: 10.1093/ecam/nen004.
- Sun Y, Liu T, Sun C, Luo Q. 2022. Chemical constituents of Plectranthus hadiensis extract and its control effect on Tetranychus kanzawai. New J Chem. 2022:1–6. doi: 10.1155/2022/5609391.
- Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. 2011. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 28(10):2731–2739. doi: 10.1093/molbev/msr121.
- Tao AE, Zhao FY, Xia CL. 2019. Characterization of the complete chloroplast genome of Ajuga forrestii (Lamiaceae), a medicinal plant in southwest of China. Mitochondrial DNA B Resour. 4(2):3969–3970. doi: 10.1080/23802359.2019.1689193.
- Wang H, Wang Y, Cheng H, Xue M, Wang J, Yue Z. 2021. The complete chloroplast genome of Schizonepeta tenuifolia (Benth.) Briq., a traditional Chinese herb. Mitochondrial DNA B Resour. 6(3):907–908. doi: 10.1080/23802359.2021.1886884.
- Wang H, Wang Y, Lu Y, Zhu S, Huang J, Yue C. 2022. Characterization of the complete chloroplast genome sequence of Isodon japonicus (N. Burman) H. Hara (Lamiaceae). Mitochondrial DNA B Resour. 7(9):1713–1715. doi: 10.1080/23802359.2022.2123718.
- Wang K, Li L, Hua Y, Zhao M, Li S, Sun H, Lv Y, Wang Y. 2017. The complete chloroplast genome of Mentha spicata, an endangered species native to South Europe. Mitochondrial DNA B Resour. 2(2):907–909. doi: 10.1080/23802359.2017.1413311.
- Yu Q, Huang C, Yang Z. 2014. Nuclear-encoded factors associated with the chloroplast transcription machinery of higher plants. Front Plant Sci. 5:316. doi: 10.3389/fpls.2014.00316.
- Yue Z, Wang Y, Zhou B, Wang H. 2021. The complete chloroplast genome of Isodon rubescens, a traditional Chinese herb. Mitochondrial DNA Part B. 6(2):337–338. doi: 10.1080/23802359.2020.1860704.
- Zhang HY, Ma WZ, Yan HF, Wang DQ. 2020. Characterization of the complete plastid genome of Chinese medicinal plant Isodon serra (Lamiaceae). Mitochondrial DNA B Resour. 5(3):2111–2112. doi: 10.1080/23802359.2020.1765429.
- Zhang HY, Xia J, Ma WZ. 2020. The complete plastome of a folk medicinal herb Isodon lophanthoides var. graciliflorus. Mitochondrial DNA B Resour. 5(3):2219–2221. doi: 10.1080/23802359.2020.1768949.
- Zhao F, Drew BT, Chen Y-P, Hu G-X, Li B, Xiang C-L. 2020. The chloroplast genome of Salvia: genomic characterization and phylogenetic analysis. Int J Plant Sci. 181(8):812–830. doi: 10.1086/710083.
- Zubair Filimban F, Yarádua SS, Bello A, Choudhry H. 2022. Complete chloroplast genomes of two mint species (Lamiaceae) from Al-Madinah, Saudi Arabia: phylogenetic and genomic comparative analyses. Mitochondrial DNA B Resour. 7(10):1797–1799. doi: 10.1080/23802359.2022.2130713.
