Abstract
Lanmaoa macrocarpa is a boletoid mushroom from the family Boletaceae and was named after its relatively larger basidiocarp and bluish color change when bruised. At present, its mitochondrial genome and phylogenetic relationships with other boletes remain unexplored. In this study, we sequenced the mitochondrial genome of L. macrocarpa using next-generation sequencing technology and found that its mitochondrial genome, a circular DNA molecule of 38,139 bp, comprised 15 core protein-coding genes, 26 transfer RNA genes and two ribosomal RNA genes. The mitochondrial genome had a base composition of A (37.05%), C (12.08%), G (11.42%) and T (39.45%) with a GC content of 23.50%. A phylogenetic tree based on 20 mitochondrial genomes was constructed, which revealed the phylogenetic relationships of this species with related boletes for the first time.
Introduction
Lanmaoa macrocarpa Zeng, Chai & Jiang 2019, a member of the boletoid mushroom, got is name for its relatively larger basidiocarps. In addition, it’s also distinguished by immediate bluish color change when bruised and mild taste (Chai et al. Citation2019). The basidiocarp of L. macrocarpa was shown in . Lanmaoa macrocarpa has long been consumed in its collection site, Jiangxi Province, whereas the complete mitochondrial genome of this edible mushroom has not been sequenced yet. In this study, the complete mitochondrial genome of the East Asia bolete was sequenced on an Illumina HiSeq 2500 Platform, and the phylogenetic relationships with related species were also analyzed.
Materials
The specimen of L. macrocarpa was collected from Jiulingshan National Nature Reserve, Jiangxi Province, China (115°21′09″E, 28°54′43″N) and deposited in the Cryptogamic Herbarium, Kunming Institute of Botany, Chinese Academy of Sciences (http://groups.kib.cas.cn/kun/) under the voucher number KUN-HKAS 105258 (contact Kuan Zhao, [email protected]). This specimen was identified by the corresponding author. Specific permission is not needed as no endangered or protected species were involved.
Methods
The total DNA was extracted from the context of the basidiocarp using the CTAB method (Doyle and Doyle Citation1987) and then sequenced by Sangon Biotech Co., Ltd (Shanghai, China) for clean reads. For assembly with GetOrganelle (Jin et al. Citation2020), we used the fungus database (-F fungus_mt) to identify, filter, and assemble target-associated reads. The annotation was completed by MITOS Web Server using the genetic code 4 (Donath et al. Citation2019) and the annotated protein-coding genes (PCGs) were then further refined using the open reading frame (ORF) finder from the National Center for Biotechnology Information (NCBI, https://www.ncbi.nlm.nih.gov/orffinder/). tRNA genes were also identified using tRNAscan-SE v1.3.1 (Lowe & Chan Citation2016). Intron types were verified through RNAweasel v5.2.1 (Lang et al. Citation2007). OGDRAW (Greiner et al. Citation2019) was applied to visualize the gene map.
A total of 19 mitochondrial genomes were downloaded from NCBI and the Joint Genome Institute (JGI, https://mycocosm.jgi.doe.gov/mycocosm/home) database, including sixteen from the family Boletaceae, with two species from the genus Paxillus (Paxillaceae, Boletales) and one species from Suillus (Suillaceae, Boletales) as outgroups. 15 core PCGs were extracted and aligned respectively by MAFFT v7.037 (Katoh et al. Citation2019), then the individual alignments were concatenated using Phyutility (Smith and Dunn Citation2008). The final concatenated alignment was analyzed using MrBayes v3.2.6 and RAxML v 8.0.0 for BI and ML methods, respectively (Ronquist and Huelsenbeck Citation2003; Stamatakis Citation2006). BI analyses were conducted by setting generations to one million and runs were automatically terminated once the average standard deviation of split frequencies went below 0.01. Other parameters were kept at their default settings. In the ML analyses, bootstrap values (BS) were assessed through an ultrafast bootstrap approach with 1000 replicates.
Results
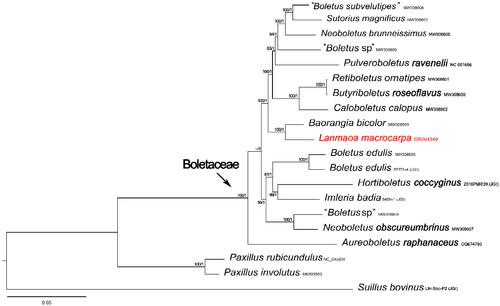
The complete mitochondrial genome sequence of L. macrocarpa (GenBank accession no. OR004349) was found to be 38,139 bp in length, which is close to that of other boletes, ranging from 32, 883 bp to 48, 298 bp (Li et al. Citation2021). The gene map of L. macrocarpa was shown in . The complete mitochondrial genome comprised 15 core PCGs (atp6, atp8, atp9, cob, cox1, cox2, cox3, nad1, nad2, nad3, nad4, nad4L, nad5, nad6 and rps3), 26 transfer RNA genes and two ribosomal RNA genes. The mitochondrial genome had a base composition of A (37.05%), C (12.08%), G (11.42%) and T (39.45%) with a GC content of 23.50%. The start-codon of all the 15 PCGs is ATG and the termination codon for 14 PCGs is TAA. The atp8 gene is the only exception, as it has a stop codon of TAG. Additionally, an intron belonging to group IA was identified in the rrnL gene. The phylogenetic result discovered that the newly sequenced L. macrocarpa was sister to Baorangia bicolor as shown in . In addition, the relationships among the boletoid mushrooms were also indicated.
Figure 2. The mitochondrial genome map of Lanmaoa macrocarpa. Genes outside and inside the outer circle are transcribed in counterclockwise and clockwise directions, respectively. Intron-contained genes were labeled with *. GC and at contents across the genome are shown with dark and light shading, respectively, inside the inner circle.
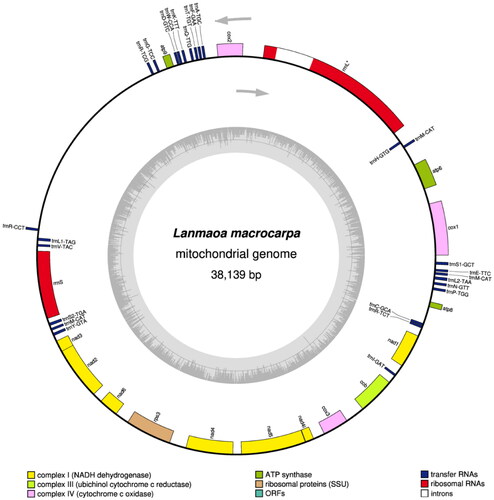
Figure 3. Phylogenetic tree of Lanmaoa macrocarpa and related taxa based on Bayesian inference (BI) and maximum likelihood (ML) analyses of 15 core protein coding genes. The GenBank accession numbers from NCBI or the information of voucher specimens from JGI followed the species names. The newly sequenced mitogenome is marked in red. Numbers near the nodes indicate bootstrap support values (>50%) and posterior probabilities (>0.95). the scale bar refers to 0.05 nucleotide substitutions per character.
Discussion and conclusion
This study firstly reported the whole mitochondrial genome of an edible bolete Lanmaoa macrocarpa. In addition, the phylogenic analyses results enriched and supplemented the previous studies on the family Boletaceae (Li et al. Citation2021; Cho et al. Citation2022). Bolete is important macro-fungi group for its ecological and economical value. As it is difficult to distinguish the boletes just from morphological features, several molecular markers have been selected and widely used to identify them (Wu et al. Citation2016; Zhao et al. Citation2020). As reported in previous study, mitochondrial genome can also provide more genetic information than most of molecular markers and was proved to be a potential option to elucidate the phylogenetic relationships of boletes (Li et al. Citation2021). The family Boletaceae harbors more than 800 species around the globe but only a dozen mitochondrial genomes were sequenced and deposited in public database (NCBI and JGI), the phylogeny of Boletaceae will be elucidated deeply in the future when more mitochondrial genomes be sequenced.
Author contributions
YTZ and LLC collected the samples, performed the analysis and interpretation of the data, and prepared the manuscript. KZ devised the project, revised the manuscript critically and approved the final version to be published. All authors discussed and critically revised the results and contributed to the final version of the manuscript.
Ethical approval
No ethical issues were involved in this study. The collection of the mushroom was legal and reasonable. Information of the voucher specimen and who identified it were included in the manuscript.
Supplemental Material
Download PNG Image (222.5 KB)Disclosure statement
The authors have declared that no competing interests exist.
Data availability statement
The mitochondrial genome data is available with the accession number of OR004349 in the GenBank of NCBI (https://www.ncbi.nlm.nih.gov/). And the associated Bioproject, SRA, Bio-sample numbers are PRJNA957944, SRR24299300 and SAMN34273906, respectively.
Additional information
Funding
References
- Chai H, Liang Z-Q, Xue R, Jiang S, Luo S-H, Wang Y, Wu L-L, Tang L-P, Chen Y, Hong D, et al. 2019. New and noteworthy boletes from subtropical and tropical China. MycoKeys. 46:55–96. doi: 10.3897/mycokeys.46.31470.
- Cho S-E, Kwag Y-N, Han S-K, Lee D-H, Kim C-S. 2022. Complete mitochondrial genome sequence of Pulveroboletus ravenelii (Boletales, Basidiomycota). Mitochondrial DNA B Resour. 7(9):1581–1582. doi: 10.1080/23802359.2022.2110006.
- Donath A, Jühling F, Al-Arab M, Bernhart SH, Reinhardt F, Stadler PF, Middendorf M, Bernt M. 2019. Improved annotation of protein-coding genes boundaries in metazoan mitochondrial genomes. Nucleic Acids Res. 47(20):10543–10552. doi: 10.1093/nar/gkz833.
- Doyle J-J, Doyle J-L. 1987. A rapid DNA isolation procedure for small quantities of fresh leaf material. Phytochem Bull. 19:11–15. doi: 10.1016/0031-9422(80)85004-7.
- Greiner S, Lehwark P, Bock R. 2019. OrganellarGenomeDRAW (OGDRAW) version 1.3.1: expanded toolkit for the graphical visualization of organellar genomes. Nucleic Acids Res. 47(W1):W59–W64. doi: 10.1093/nar/gkz238.
- Jin J-J, Yu W-B, Yang J-B, Song Y, DePamphilis C-W, Yi T-S, Li D-Z. 2020. GetOrganelle: a fast and versatile toolkit for accurate de novo assembly of organelle genomes. Genome Biol. 21(1):241. doi: 10.1186/s13059-020-02154-5.
- Katoh K, Rozewicki J, Yamada K-D. 2019. MAFFT online service: multiple sequence alignment, interactive sequence choice and visualization. Brief Bioinform. 20(4):1160–1166. doi: 10.1093/bib/bbx108.
- Lang B-F, Laforest M-J, Burger G. 2007. Mitochondrial introns: a critical view. Trends Genet. 23(3):119–125. doi: 10.1016/j.tig.2007.01.006.
- Li Q, Wu P, Li L-J, Feng H-Y, Tu W-Y, Bao Z-J, Xiong C, Gui M-Y, Huang W-L. 2021. The first eleven mitochondrial genomes from the ectomycorrhizal fungal genus (Boletus) reveal intron loss and gene rearrangement. Int J Biol Macromol. 172:560–572. doi: 10.1016/j.ijbiomac.2021.01.087.
- Lowe T-M, Chan P-P. 2016. tRNAscan-SE On-line: integrating search and context for analysis of transfer RNA genes. Nucleic Acids Res. 44(W1):W54–W57. doi: 10.1093/nar/gkw413.
- Ronquist F, Huelsenbeck J-P. 2003. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 19(12):1572–1574. doi: 10.1093/bioinformatics/btg180.
- Smith S-A, Dunn C-W. 2008. Phyutility: a phyloinformatics tool for trees, alignments and molecular data. Bioinformatics. 24(5):715–716. doi: 10.1093/bioinformatics/btm619.
- Stamatakis A. 2006. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics. 22(21):2688–2690. doi: 10.1093/bioinformatics/btl446.
- Wu G, Li Y-C, Zhu X-T, Zhao K, Han L-H, Cui Y-Y, Li F, Xu J, Yang Z-L. 2016. One hundred noteworthy boletes from China. Fungal Divers. 81(1):25–188. doi: 10.1007/s13225-016-0375-8.
- Zhao K, Zhang F-M, Zeng Q-Q, Han L-H, Li Y-C. 2020. Tylopilus jiangxiensis, a new species of Tylopilus s. str. from China. Phytotaxa. 434(3):281–291. doi: 10.11646/phytotaxa.434.3.6.

