Abstract
Elymus alashanicus (Keng) S. L. Chen, a herbaceous plant endemic to China, plays a crucial role in the local ecosystems. In this study, we sequenced and characterized the complete chloroplast (cp) genome of E. alashanicus, which is 135,072 bp in length and arranged in a circular form. The cp genome includes a pair of inverted repeats (IRa and IRb) of 20,813 bp each, separated by a large single-copy (LSC) region of 80,678 bp and a small single-copy (SSC) region of 12,768 bp. The cp genome contains 130 genes, including 83 protein-coding genes, 39 tRNA genes, and eight rRNA genes. Phylogenetic analysis revealed that E. alashanicus is closely related to Elymus breviaristatus and Campeiostachys dahurica var. tangutorum in current sampling. Our findings provide valuable insights into the cp genome of E. alashanicus, which could contribute to further studies on the evolution and conservation of this species.
Introduction
Elymus alashanicus (Keng) S. L. Chen, a perennial herb of the genus Roegneria (Triticeae), was first described in 1963 and is widely distributed in northwest China (Zhang et al. Citation2009; Lei et al. Citation2016). This species is known for its excellent genes of disease and stress resistance and high feed value, making it an important natural gene bank for forage breeding and improvement of wheat-related plants (Cai Citation2002). However, there is limited information on the chloroplast (cp) genomes of E. alashanicus. An in-depth analysis of the cp genomics of this genus will provide a foundation for phylogenetic studies and aid taxonomical analyses.
Materials and methods
Sample collection and preservation
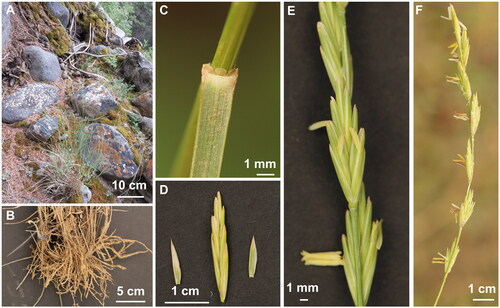
Fresh leaves of E. alashanicus were collected from Helan Mountain (N38°22′52″, E105°42′53″) in Inner Mongolia, China (). A specimen was deposited at herbarium of Inner Mongolia Normal University (NMTC, http://bio.imnu.edu.cn/info/1097/1401.htm, contact person and email: FengJin, [email protected]) under the voucher number HLS20190828-67.
DNA extraction and sequencing
Total genomic DNA was extracted using the method of Doyle and Doyle (Doyle and Doyle Citation1987). Short-insert library (insert size, 300 bp) was prepared and then sequenced using the Illumina HiSeq platform in NextOmics (Wuhan, China).
Assembly, annotation, and visualization
The complete genome was de novo assembled using NOVOPlasty (Dierckxsens et al. Citation2017) and annotated with the online annotation tool GeSeq (https://chlorobox.mpimp-golm.mpg.de/OGDraw.html). The complete plastome of E. alashanicus has been deposited in GenBank under accession number OL444890. A circular map of its plastome was visualized using the CPGView online web (http://www.1kmpg.cn/cpgview) (Liu et al. Citation2023).
Phylogenetic reconstruction
To determine the phylogenetic position of E. alashanicus, the complete cp genome sequences of 30 species of Poaceae plants, plus Arabidopsis thaliana (AP000423.1) and Solanum tuberosum (NC_008096.2) as outgroups were obtained from NCBI (https://www.ncbi.nlm.nih.gov/).
Bootstrapped maximum-likelihood (ML) with 1000 replicates was constructed based on GTR + I+GAMMA model in jModelTest v. 2.1.10. Additionally, Bayesian inference (BI) trees were constructed using the Markov chain Monte Carlo (MCMC) method in MrBayes (v3.2.7) (Darriba et al. Citation2012). Each sequence was aligned using the ‘auto’ mode in MAFFT (v7.427), then the algorithm was run for 1,000,000 iterations, and samples were taken every 100 iterations (Katoh and Standley Citation2013). The first 25% of the resulting trees were removed as ‘burn-in’, and the majority-rule consensus tree was used as the final result.
Results
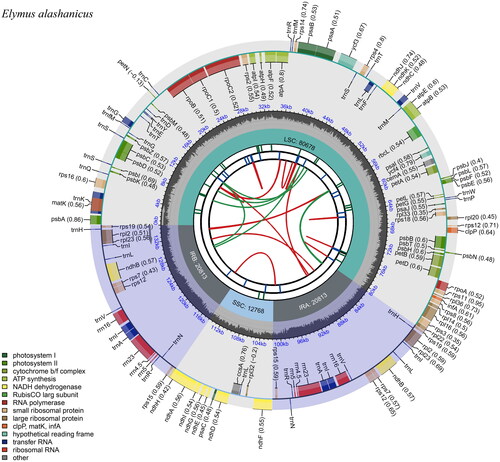
The cp genome of E. alashanicus is 135,072 bp in length, with an average sequencing depth of 466X (Supplementary Figure S1). It exhibited a typical four-stage structure, consisting of a large single-copy (LSC) region of 80,678 bp, a small single-copy (SSC) region of 12,768 bp, and two inverted repeat regions (IRa/IRb), both measuring 20,813 bp (). The content of CG in whole cp genome was 38.33%, and a total of 130 genes were encoded, including 83 protein-coding genes, 39 tRNA genes, and eight rRNA genes. Among them, 10 protein-coding genes (rps16, atpF, petB, petD, rpl16, rpl2, ndhB, ndhA, ndhB, and rpl2) and eight tRNA genes (trnK-UUU, trnG-UCC, trnL-UAA, trnV-UAC, trnI-GAU, trnA-UGC, trnA-UGC, and trnI-GAU) containing one intron and three genes (ycf3, rps12, and rps12) had two introns. Furthermore, a trans-spliced gene rps12 and three small-exon genes (petB, petD, and rpl16) were verified and annotated using multiple sequence alignment (Supplementary Figures S2 and S3).
Figure 2. Schematic circular map of overall features of E. alashanicus chloroplast genome. Graphic showing features of its plastome was generated using CPGview. The map contains six tracks. From the inner circle, the first track depicts the dispersed repeats connected by red (forward direction) and green (reverse direction) arcs, respectively. The second track shows the long tandem repeats as short blue bars. The third track displays the short tandem repeats or microsatellite sequences as short bars with different colors. The fourth track depicts the sizes of the inverted repeats (IRa and IRb), small single-copy (SSC), and large single-copy (LSC). The fifth track plots the distribution of GC contents along the plastome. The sixth track displays the genes belonging to different functional groups with different colored boxes. The outer and inner genes are transcribed in the clockwise and counterclockwise directions, respectively.
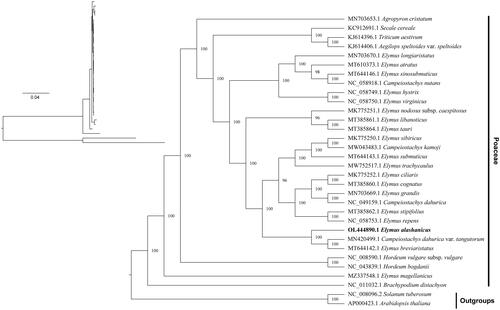
Phylogenetic analysis based on the whole cp genome sequences of 31 reported species and the new data from this study showed that E. alashanicus was clustered with Elymus breviaristatus and Campeiostachys dahurica var. tangutorum with full supportive values of the BI trees ().
Figure 3. Maximum-likelihood and Bayesian inference phylogenetic trees of 30 Poaceae species with Arabidopsis thaliana and Solanum tuberosum as outgroups. The main phylogenetic tree was presented based on the BI tree, because of highly similarity among the sequences, showed lower branch support of the ML evolutionary tree. Species names are displayed with indicate lines in the right side, and the numbers above branches indicate supportive values of BI phylogenetic trees. GenBank accession numbers of the following sequences were used to construct the phylogenetic tree: Agropyron cristatum MN703653.1 (Chen et al. Citation2018), Secale cereale KC912691.1 (Li et al. Citation2015), Triticum aestivum KJ614396.1 (Han et al. Citation2021), Aegilops speltoides var. speltoides KJ614406.1 (Chen et al. Citation2018), Elymus longiaristatus MN703670.1 (Hu et al. Citation2015), Elymus atratus MT610373.1 (Gao et al. Citation2014), Elymus sinosubmuticus MT644146.1 (Tan et al. Citation2022), Campeiostachys nutans NC_058918.1 (Dong et al. Citation2015), Elymus hystrix NC_058749.1 (Dong et al. Citation2015), Elymus virginicus NC_058750.1 (Dong et al. Citation2015), Elymus nodosus subsp. caespitosus MK775251.1 (Chen et al. Citation2020), Elymus libanoticus MT385861.1 (Xia and Liu Citation2020), Elymus tauri MT385864.1 (Liu et al. Citation2021), Elymus sibiricus MK775250.1 (Chen et al. Citation2022), Campeiostachys kamoji MW043483.1 (Liu et al. Citation2021), Elymus submuticus MT644143.1 (Y. NI 2011), Elymus trachycaulus MW752517.1 (Wu et al. Citation2016), Elymus ciliaris MK775252.1 (Hu et al. Citation2013), Elymus cognatus MT385860.1 (Hu et al. Citation2015), Elymus grandis MN703669.1 (Yu et al. Citation2010), Campeiostachys dahurica NC_049159.1 (Tan et al. Citation2021), Elymus stipifolius MT385862.1 (Yang et al. Citation2017), Elymus repens NC_058753.1 (Mason-Gamer Citation2008), Campeiostachys dahurica var. tangutorum MN420499.1 (Yang et al. Citation2015; Jing et al. Citation2019), Elymus breviaristatus MT644142.1 (Tan et al. Citation2022), Hordeum vulgare subsp. vulgare NC_008590.1 (Zhelyazkova et al. Citation2012), Hordeum bogdanii NC_043839.1 (Cui et al. Citation2021), Elymus magellanicus MZ337548.1 (Wu et al. Citation2022), Brachypodium distachyon NC_011032.1 (Sancho et al. Citation2018), Solanum tuberosum NC_008096.2 (Occhialini et al. Citation2020), and Arabidopsis thaliana AP000423.1 (Provan Citation2000).
Discussion and conclusions
E. alashanicus is a species with excellent production performance and high economic value due to its strong tillering ability and resistance to poverty and drought. This study presented the first complete cp genome of E. alashanicus and annotated its structure. The phylogenetic analysis revealed that E. alashanicus was closely related to Elymus breviaristatus and Campeiostachys dahurica var. tangutorum. E. alashanicus is also known as Roegneria alashanicus (https://www.worldfloraonline.org/taxon/wfo-0000895903), and the classification of plants from these genera has always been inconsistent due to their complex morphology. Here, the complete cp genome sequence of E. alashanicus suggests that it is genetically more related to the genus Elymus. This study could provide valuable genomic information for further investigation of its taxonomy evolution and population genetics.
Ethical approval
No specific permissions were needed to perform this research as E. alashanicus is not a protected plant, and no damage could be caused to its population.
Author contributions
Feng Jin designed the project and performed the sample collection. Bao Sarina and Jinghuan Li analyzed and interpretated the sequencing data and drafted the paper. Feng Jin revised it and approved the final version to be published. Sarina and Feng Jin contributed equally to this work and all authors agree to be accountable for all aspects of the work.
Supplemental Material
Download TIFF Image (333.9 KB)Supplemental Material
Download TIFF Image (800.4 KB)Supplemental Material
Download TIFF Image (757.1 KB)Disclosure statement
No potential conflict of interest was reported by the authors. All authors revised and approved the manuscript.
Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI at https://www.ncbi.nlm.nih.gov/ under the accession no. OL444890.1. The associated BioProject, SRA, and Bio-Sample numbers are PRJNA778469, SRR16913047, and SAMN22960735, respectively.
Additional information
Funding
References
- Cai L. 2002. Geographical distribution of Roegneria C. Koch (Poaceae). Acta Bot Bor Occid Sin. 22(4):913–923.
- Chen C, Zheng Z, Wu D, Tan L, Yang C, Liu S, Lu J, Cheng Y, Sha L, Wang Y, et al. 2022. Morphological, cytological, and molecular evidences for natural hybridization between Roegneria stricta and Roegneria turczaninovii (Triticeae: Poaceae). Ecol Evol. 12(1):e8517.
- Chen N, Chen W-J, Yan H, Wang Y, Kang H-Y, Zhang H-Q, Zhou Y-H, Sun G-L, Sha L-N, Fan X, et al. 2020. Evolutionary patterns of plastome uncover diploid–polyploid maternal relationships in Triticeae. Mol Phylogenet Evol. 149:106838. doi:10.1016/j.ympev.2020.106838.
- Chen N, Sha L-N, Dong Z-Z, Tang C, Wang Y, Kang H-Y, Zhang H-Q, Yan X-B, Zhou Y-H, Fan X, et al. 2018. Complete structure and variation of the chloroplast genome of Agropyron cristatum (L.) Gaertn. Gene. 640:86–96. doi:10.1016/j.gene.2017.10.009.
- Cui G, Wang C, Wei X, Wang H, Wang X, Zhu X, Li J, Yang H, Duan H. 2021. Complete chloroplast genome of Hordeum brevisubulatum: genome organization, synonymous codon usage, phylogenetic relationships, and comparative structure analysis. PLOS One. 16(12):e0261196. doi:10.1371/journal.pone.0261196.
- Darriba D, Taboada GL, Doallo R, Posada D. 2012. jModelTest 2: more models, new heuristics and parallel computing. Nat Methods. 9(8):772–772. doi:10.1038/nmeth.2109.
- Dierckxsens N, Mardulyn P, Smits G. 2017. NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 45(4):e18. doi:10.1093/nar/gkw955.
- Dong Z-Z, Fan X, Sha L-N, Wang Y, Zeng J, Kang H-Y, Zhang H-Q, Wang X-L, Zhang L, Ding C-B, et al. 2015. Phylogeny and differentiation of the St genome in Elymus L. sensu lato (Triticeae; Poaceae) based on one nuclear DNA and two chloroplast genes. BMC Plant Biol. 15(1):179. doi:10.1186/s12870-015-0517-2.
- Doyle JJ, Doyle JL. 1987. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull. 19:11–15.
- Gao G, Gou X, Wang Q, Zhang Y, Deng J, Ding C, Zhang L, Zhou Y, Yang R. 2014. Phylogenetic relationships and Y genome origin in Chinese Elymus (Triticeae: Poaceae) based on single copy gene DMC1. Biochem Syst Ecol. 57:420–426. doi:10.1016/j.bse.2014.09.019.
- Han Y, Gao Y, Zhou H, Zhai X, Ding Q, Ma L. 2021. Mitochondrial genes are involved in the fertility transformation of the thermosensitive male-sterile line YS3038 in wheat. Mol Breeding. 41(10):61. doi:10.1007/s11032-021-01252-x.
- Hu Q, Sun D, Sun G. 2015. Molecular phylogeny revealed distinct origin of the Y and St genome in Elymus longearistatus (Triticeae: Poaceae). Mol Phylogenet Evol. 85:141–149. doi:10.1016/j.ympev.2015.02.012.
- Hu Q, Yan C, Sun G. 2013. Phylogenetic analysis revealed reticulate evolution of allotetraploid Elymus ciliaris. Mol Phylogenet Evol. 69(3):805–813. doi:10.1016/j.ympev.2013.06.023.
- Jing M, Ma Y, Li H, Wang J. 2019. Characterization of the complete plastome of Elymus tangutorum (Poaceae: Triticeae). Mitochondrial DNA B Resour. 4(2):3356–3357. doi:10.1080/23802359.2019.1674217.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780. doi:10.1093/molbev/mst010.
- Lei Y-X, Zhang Y, Li Y-y, Fan X, Sha L-N, Wang Y, Kang H-Y, Zhou Y-H, Zhang H-Q. 2016. Phylogenetic analysis of the species with awnless lemma in Roegneria (Poaceae, Triticeae) based on single copy of nuclear gene DMC1. Biochem Syst Ecol. 65:185–191. doi:10.1016/j.bse.2016.02.021.
- Li LF, Liu B, Olsen KM, Wendel JF. 2015. A re-evaluation of the homoploid hybrid origin of Aegilops tauschii, the donor of the wheat D-subgenome. New Phytol. 208(1):4–8. doi:10.1111/nph.13294.
- Liu M, Zhang Y, Zhang Y, Cui G, Zhou Q, Wei X. 2021. Characterization of the complete chloroplast genome of Elymus kamoji (Ohwi) S. L. Chen. Mitochondrial DNA B Resour. 6(11):3177–3178. doi:10.1080/23802359.2021.1882898.
- Liu S, Ni Y, Li J, Zhang X, Yang H, Chen H, Liu C. 2023. CPGView: a package for visualizing detailed chloroplast genome structures. Mol Ecol Resour. 23(3):694–704. doi:10.1111/1755-0998.13729.
- Mason-Gamer RJ. 2008. Allohexaploidy, introgression, and the complex phylogenetic history of Elymus repens (Poaceae). Mol Phylogenet Evol. 47(2):598–611. doi:10.1016/j.ympev.2008.02.008.
- Ni Y, Asamoah-Odei N, Sun G. 2011. Maternal origin, genome constitution and evolutionary relationships of polyploid Elymus species and Hordelymus europaeus. Biol Plant. 55(1):68–74. doi:10.1007/s10535-011-0009-7.
- Occhialini A, Pfotenhauer AC, Frazier TP, Li L, Harbison SA, Lail AJ, Mebane Z, Piatek AA, Rigoulot SB, Daniell H, et al. 2020. Generation, analysis, and transformation of macro-chloroplast Potato (Solanum tuberosum) lines for chloroplast biotechnology. Sci Rep. 10(1):21144. doi:10.1038/s41598-020-78237-x.
- Provan J. 2000. Novel chloroplast microsatellites reveal cytoplasmic variation in Arabidopsis thaliana. Mol Ecol. 9(12):2183–2185. doi:10.1046/j.1365-294X.2000.105316.x.
- Sancho R, Cantalapiedra CP, López-Alvarez D, Gordon SP, Vogel JP, Catalán P, Contreras-Moreira B. 2018. Comparative plastome genomics and phylogenomics of Brachypodium: flowering time signatures, introgression and recombination in recently diverged ecotypes. New Phytol. 218(4):1631–1644. doi:10.1111/nph.14926.
- Tan L, Huang Q-X, Song Y, Wu D-D, Cheng Y-R, Zhang C-B, Sha L-N, Fan X, Kang H-Y, Wang Y, et al. 2022. Biosystematics studies on Elymus breviaristatus and Elymus sinosubmuticus (Poaceae: Triticeae). BMC Plant Biol. 22(1):57. doi:10.1186/s12870-022-03441-y.
- Tan L, Zhang H-Q, Chen W-H, Deng M-Q, Sha L-N, Fan X, Kang H-Y, Wang Y, Wu D-D, Zhou Y-H. 2021. Genome composition and taxonomic revision of Elymus purpuraristatus and Roegneria calcicola (Poaceae: Triticeae) based on cytogenetic and phylogenetic analyses. Bot J Linn Soc. 196(2):242–255. doi:10.1093/botlinnean/boaa103.
- Wu D-C, He D-M, Gu H-L, Wu P-P, Yi X, Wang W-J, Shi H-F, Wu D-X, Sun G. 2016. Origin and evolution of allopolyploid wheatgrass Elymus fibrosus (Schrenk) Tzvelev (Poaceae: Triticeae) reveals the effect of its origination on genetic diversity. PLOS One. 11(12):e0167795. doi:10.1371/journal.pone.0167795.
- Wu X, Chen X, Huang Z, Ren C, Hu T, Ru Z. 2022. De novo assembly and characterization of the complete chloroplast genome of Elymus magellanicus (E.Desv.) A.Love, 1984 (Poaceae, Pooideae). Mitochondrial DNA B Resour. 7(10):1873–1875. doi:10.1080/23802359.2022.2135400.
- Xia M, Liu R. 2020. Characterization of the complete chloroplast genome of Elymus dahuricus. Mitochondrial DNA Part B. 5(2):1512–1513. doi:10.1080/23802359.2020.1742218.
- Yang C-R, Zhang H-Q, Zhao F-Q, Liu X-Y, Fan X, Sha L-N, Kang H-Y, Wang Y, Zhou Y-H. 2015. Genome constitution of Elymus tangutorum (Poaceae: Triticeae) inferred from meiotic pairing behavior and genomic in situ hybridization. J Syst Evol. 53(6):529–534. doi:10.1111/jse.12155.
- Yang Y, Fan X, Wang L, Zhang H-Q, Sha L-N, Wang Y, Kang H-Y, Zeng J, Yu X-F, Zhou Y-H. 2017. Phylogeny and maternal donors of Elytrigia Desv. sensu lato (Triticeae; Poaceae) inferred from nuclear internal-transcribed spacer and trnL-F sequences. BMC Plant Biol. 17(1):207. doi:10.1186/s12870-017-1163-7.
- Yu H-Q, Zhang C, Ding C-B, Zhang H-Q, Zhou Y-H. 2010. Genome constitutions of Roegneria alashanica, R. elytrigioides, R. magnicaespes and R. grandis (Poaceae: Triticeae) via genomic in-situ hybridization. Nord J Bot. 28(2):206–211. doi:10.1111/j.1756-1051.2009.00611.x.
- Zhang C, Fan X, Yu H-Q, Zhang H-Q, Wang X-L, Zhou Y-H. 2009. Phylogenetic analysis of questionable tetraploid species in Roegneria and Pseudoroegneria (Poaceae: Triticeae) inferred from a gene encoding plastid acety1-CoA carboxylase. Biochem Syst Ecol. 37(4):412–420. doi:10.1016/j.bse.2009.04.011.
- Zhelyazkova P, Sharma CM, Förstner KU, Liere K, Vogel J, Börner T. 2012. The primary transcriptome of barley chloroplasts: numerous noncoding RNAs and the dominating role of the plastid-encoded RNA polymerase. Plant Cell. 24(1):123–136. doi:10.1105/tpc.111.089441.