Abstract
Auricularia delicata (Mont.) Henn. 1893 is an edible and medicinal jelly mushroom popular in China. Here, we report the assembly and annotation of a complete A. delicata mitochondrial genome based on data sequenced using an Illumina NovaSeq 6000 platform. The length of the complete circular A. delicata mitochondrial genome is 189,696 bp, with a GC content of 34.1%. The A. delicata mitochondrial genome contains 60 genes, including 32 protein-coding genes, 26 tRNA genes, and two rRNA genes. Phylogenetic analysis indicated that A. delicata clustered with the Auricularia group, alongside A. auricula-judae and A. heimuer. Additionally, A. delicata was found to be genetically distant from other species of Polyporales, Russulales, and Agaricales. This genome will provide an invaluable reference for the continued study and utilization of A. delicata and other Auricularia species.
1. Introduction
Auricularia delicata (Mont.) Henn. 1893 is a jelly mushroom utilized as a source of food and medicine across certain regions of Africa and eastern Asia, and particularly in China and India (Wangkheirakpam et al. Citation2018; Li et al. Citation2020; Muharagi et al. Citation2020). A. delicata grows on fresh-cut wood, decaying logs, and tree trunks across several temperate, tropical, and subtropical areas (Looney et al. Citation2013). A. delicata belongs to a species complex which can be roughly divided between American and Australian groups (Looney et al. Citation2013). Through advancements in fungal cultivation techniques, A. delicata is being successfully cultivated in China and other countries (Qian et al. Citation2020). To address gaps in our understanding of the phylogeny and evolutionary history of this economically important fungal species, here, we report the first complete A. delicata mitochondrial genome. This genome will provide an invaluable reference for the continued study and utilization of A. delicata and other Auricularia species.
2. Materials and methods
2.1. Materials
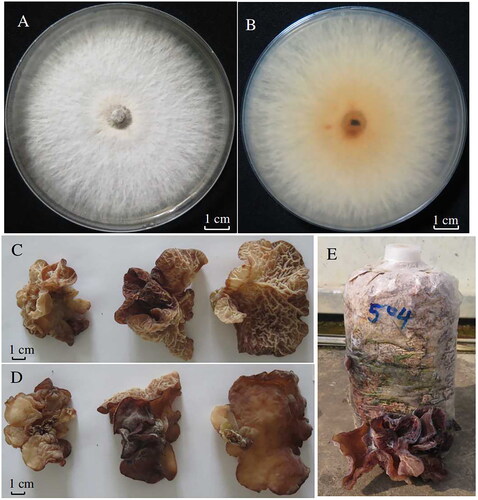
An A. delicata specimen found growing on a decaying stump was collected from Nanning, Guangxi Province, China (108°15′15.2604″N; 22°51′4.3272″E). A voucher specimen (No. 2018051504) was stored at the Guangxi Academy of Agricultural Sciences (GXAAS) (Wang Xiaoguo, [email protected]). The species identity of the specimen was verified through morphological investigation of the fruiting body as well as internal transcribed spacer (ITS) sequencing ().
Figure 1. Photos of the A. delicata specimen used in this study. (A) A topside view of the A. delicata mycelium; (B) an underside view of the A. delicata mycelium; (C) an underside view of the A. delicata fruiting body; (D) a topside view of the A. delicata fruiting body; (E) cultivation of A. delicata in a grow bag. Although the appearance of A. delicata fruiting bodies can vary significantly, they are generally large, dark purple-brown, soft, floppy, and bowl-shaped. The underside of the A. delicata fruiting body is porous, which differentiates this species from other Auricularia species. All photographs were taken by Dr. Wang Xiaoguo with a Canon EOS M50 camera in a laboratory in Nanning, China (108°14′ 37″ N; 22°50′ 51″ E).
2.2. DNA extraction and sequencing, and genome assembly and annotation
Total genomic DNA was extracted with a QIAamp DNA Mini Kit (Qiagen, Hilden, Germany). Sequencing was performed with an Illumina NovaSeq 6000 platform (Illumina, San Diego, CA). The library was constructed with a Nextera XT DNA Library Prep Kit (Illumina, San Diego, CA), with an average read length of 350 bp. A total of 2.63 Gb of raw sequencing reads were cleaned and edited with the NGS QC Tool Kit (Patel and Jain Citation2012). After quality control, a total of 2.61 Gb of clean reads, with a >100-fold coverage depth, were utilized for further analyses (Figure S1). The clean data exhibited a GC content of 58.48%, a Q20 of 97.81%, and a Q30 of 93.85%, indicating high quality sequencing and assembly. The high-quality reads were assembled into a complete A. delicata mitochondrial genome with the SPAdes v.3.11.0 de novo assembler (Bankevich et al. Citation2012). The complete A. delicata mitochondrial genome was annotated using MITOS (Bernt et al. Citation2013). A circular gene map of the A. delicata mitochondrial genome was created with Organellar Genome DRAW v1.3.1 (Greiner et al. Citation2019).
2.3. Phylogenetic analysis
We obtained 24 complete Agaricomycetes mitochondrial genomes by searching the National Center for Biotechnology Information (NCBI) nonredundant (nr) database. A total of 14 homologous protein-coding genes (PCGs) from each sequence were selected for comparison. The A. delicata mitochondrial genome was aligned with MAFFT 7.037 (Katoh and Standley Citation2013). The phylogenetic estimation model (GTR + F+R2) was selected using ModelFinder. A maximum-likelihood (ML) tree was constructed with IQtree 1.6 (Trifinopoulos et al. Citation2016), with Cantharellus cibarius (KC573037) used as the outgroup. Finally, the phylogenetic tree was constructed with MrBayes (Ronquist et al. Citation2012).
3. Results
3.1. Characteristics of the A. delicata mitochondrial genome
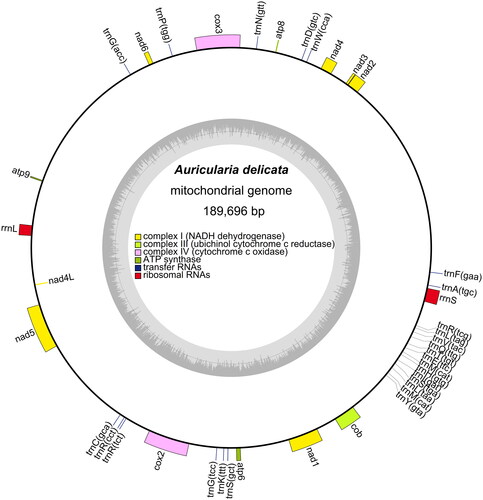
The complete, circular A. delicata mitochondrial genome (GenBank accession number OM995805) was found to be 189,696 bp in length, with a GC content of 34.1% and a base composition of 32.0% A, 33.9% T, 17.4% G, and 16.7% C. The A. delicata mitochondrial genome was found to contain 60 genes, including 32 PCGs (53.3%), 26 tRNA genes (43.3%), and two rRNA genes (rrnS and rrnL) (3.3%). The 32 conserved PCGs were found to encode apocytochrome b (cob), three ATP synthase subunits (atp6, atp8, and atp9), three cytochrome oxidase subunits (cox1, cox2, and cox3), and seven NADH: ubiquinone reductase subunits (nad1, nad2, nad3, nad4, nad4L, nad5, and nad6). The circular A. delicata mitochondrial genome map is shown in , with different colored blocks representing different categories of genes. Blocks outside the ring represent forward strand genes and blocks inside the ring represent reverse strand genes. The majority of PCGs, tRNA genes, and rRNA genes are encoded on the forward strand, with the exception of nad4L. The inner grayscale circle indicates the GC content, with the center line representing the 50% threshold.
Figure 2. Circular map of the A. delicata mitochondrial genome. Different colored blocks represent different categories of genes (listed in center). Blocks outside the ring represent forward strand genes and blocks inside the ring represent reverse strand genes. The inner grayscale circle indicates the GC content, with the center line representing the 50% threshold.
3.2. Phylogeny of A. delicata
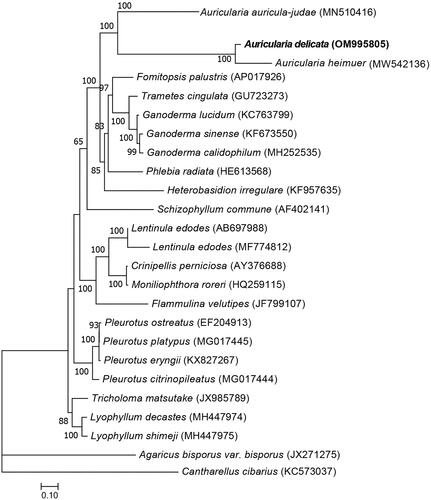
We produced a ML phylogenetic tree based on a comparison of 14 core PCGs encoded in the mitochondrial genomic sequences of A. delicata and 24 other Agaricomycetes (). As expected, A. delicata clustered with the Auricularia group with 100% bootstrap values, alongside A. auricula-judae (MN510416) and A. heimuer (MW542136.1) (Fang et al. Citation2019). Interestingly, the A. delicata mitochondrial genome was considerably larger (189,696 bp) than that of A. auricula-judae (40,586 bp) and smaller than that of A. heimuer (209,153 bp) (Fang et al. Citation2019). Additionally, A. delicata was found to be genetically distant from other species of Polyporales, Russulales, and Agaricales.
Figure 3. Phylogenetic tree showing the evolutionary relationships between A. delicata and 24 other Agaricomycetes species, with Cantharellus cibarius used as the outgroup. The tree was produced using the ML method (IQ tree 1.6) to compare 14 homologous PCGs encoded in the mitochondrial genomic sequences of each compared species. Node values indicate bootstrap support (1000 replicates).
4. Discussion and conclusions
At present, there are few published Auricularia mitochondrial genome assemblies available for study. The complete A. delicata mitochondrial genome reported here adds to the number of Auricularia mitochondrial genomes available for future work. Additionally, our contributed genome will help resolve the phylogeny of Auricularia through comparison with a previously published analysis of Auricularia rDNA ITS barcode sequences, which included A. delicata and A. auricula-judae (Looney et al. Citation2013). This genome will provide an invaluable reference for the continued study and utilization of A. delicata and other Auricularia species.
Author contributions
Xiao-guo Wang participated in the design of this study and drafted the manuscript; Shi-yan Wei and Liang-liang Qi both collected and isolated Auricularia delicata specimen and provided assistance for data acquisition and analysis; Zai-feng Yang, Jun Tang, and Zeng-liang Liu provided assistance for data acquisition, data analysis, and statistical analysis; Sheng-jin Wu provided interpretation of data collection for the work and was responsible for ensuring that the descriptions were accurate and agreed upon by all authors. All authors have read and approved the content of this manuscript.
Ethics statement
Specimen collection conformed to international ethical requirements, and did not cause damage to the local environment. Both the process and purpose of this experimental research were in line with the rules and regulations of our institute. There were no ethical issues or other conflicts of interest related to this study.
Supplemental Material
Download JPEG Image (132.1 KB)Acknowledgements
The authors would like to thank TopEdit (www.topeditsci.com) for its linguistic assistance during the preparation of this manuscript.
Disclosure statement
No potential conflicts of interest were reported by any authors.
Data availability statement
The mitochondrial genome sequence data that support the findings of this study are openly available in the NCBI (https://www.ncbi.nlm.nih.gov) GenBank under the accession number OM995805. The associated BioProject, SRA, and Bio-Sample numbers are PRJNA805016, SRR17966153, and SAMN25819720, respectively.
Additional information
Funding
References
- Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, et al. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 19(5):455–477. doi:10.1089/cmb.2012.0021.
- Bernt M, Donath A, Juhling F, Externbrink F, Florentz C, Fritzsch G, Putz J, Middendorf M, Stadler PF. 2013. MITOS: improved de novo metazoan mitochondrial genome annotation. Mol Phylogenet Evol. 69(2):313–319. doi:10.1016/j.ympev.2012.08.023.
- Fang M, Yao F, Lu L, Zhang Y, Wang P, Lu J, Wang W, Chen X. 2019. Complete mitochondrial sequence of Auricularia heimuer, one of the most popular edible fungus in China. Mitochondrial DNA B Resour. 4(2):4029–4030. doi: 10.1080/23802359.2019.1688717.
- Greiner S, Lehwark P, Bock R. 2019. Organellar Genome DRAW (OGDRAW) version 1.3.1: expanded toolkit for the graphical visualization of organellar genomes. Nucleic Acids Res. 47(W1):W59–W64. doi:10.1093/nar/gkz238.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780. doi:10.1093/molbev/mst010.
- Li X, Muharagi MS, Zhang B, Xu A. 2020. Research progress on Auricularia delicata. J Adv Biol Biot. 23(10):8–32.
- Looney BP, Joshua MP, Brandon M. 2013. Systematics of the genus Auricularia with an emphasis on species from the southeastern United States. N Am Fungi. 8(6):1–25. doi:10.2509/naf2013.008.006.
- Muharagi MS, Li X, Mabagala FS, Xu A. 2020. Studies on optimization of culture conditions and medium components for the production of mycelial biomass of Auricularia delicata under submerged fermentation. Asian J Biol. 10(4):56–67.
- Patel RK, Jain M. 2012. NGS QC Toolkit: a toolkit for quality control of next generation sequencing data. PLOS One. 7(2):e30619. doi:10.1371/journal.pone.0030619.
- Qian K, Xu A, Yang D, Li X. 2020. Biological characteristics and cultivation of a new variety of Auricularia delicata. Acta Edulis Fungi. 27(1):36–41.
- Ronquist F, Teslenko M, van der Mark P, Ayres DL, Darling A, Höhna S, Larget B, Liu L, Suchard MA, Huelsenbeck JP. 2012. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol. 61(3):539–542. doi:10.1093/sysbio/sys029.
- Trifinopoulos J, Nguyen LT, von Haeseler A, Minh BQ. 2016. W-IQ-TREE: a fast online phylogenetic tool for maximum likelihood analysis. Nucleic Acids Res. 44(W1):W232–W235. doi:10.1093/nar/gkw256.
- Wangkheirakpam SD, Joshi DD, Leishangthem GD, Biswas D, Deb L. 2018. Hepatoprotective effect of Auricularia delicata (Agaricomycetes) from India in rats: biochemical and histopathological studies and antimicrobial activity. Int J Med Mushrooms. 20(3):213–225. doi:10.1615/IntJMedMushrooms.2018025886.
