Abstract
Oreocharis argyreia var. angustifolia of Gesneriaceae is widely distributed in South China, including Guangdong, Guangxi, Hunan, and Jiangxi provinces. However, genetic information of this species is limited, further contributing to the taxonomic complications surrounding this species. Thus, in this study, we assembled and characterized the complete chloroplast genome of O. argyreia var. angustifolia as a genomic resource for future studies. The complete plastid genome was 154,675 bp in size, with a pair of inverted repeat regions of 25,329 bp each, separating the 85,977-bp large and 18,040-bp small single copy regions. A total of 131 genes were predicted, consisting of 86 protein-coding, 37 tRNA, and eight rRNA genes. The overall GC content was 37.6%. Phylogenetic analysis based on 79 shared unique CDS resulted in a fully resolved phylogenetic tree using both the maximum likelihood and Bayesian inference methods. Based on current circumscription, both methods indicated that Oreocharis is monophyletic; O. argyreia var. angustifolia diverged after O. chienii, which then followed by the divergence of the other three species included namely, O. continifolia, O. esquirolii, and O. mileensis. The genomic data obtained will be useful for future studies on the phylogenetics and evolution of Gesneriaceae.
Introduction
Oreocharis of the family Gesneriaceae has been recorded to be made up of approximately 139 species around the world (POWO Plants of the World Online Citation2022), while most of them are endemic to China (Xu et al. Citation2017). Similar to orchids, members of Gesneriaceae, including Oreocharis, are considered one of the important plant groups for evaluating local ecological environments, and therefore are of significant scientific and economic value (Li et al. Citation2018). Oreocharis argyreia var. angustifolia Pan 1987, an endemic herb native to China, was first found in the Guangxi, China (Wang et al. Citation1990). Lately, it has been discovered to grow along the border of southeast Hunan, southwest Jiangxi provinces, as well as Ruyuan of Guangdong (Fan et al. Citation2014; Mou et al. Citation2019; http://gccc.gcib.cn).
Oreocharis argyreia var. angustifolia comes with an attractive light bluish violet flower (). When compared to O. argyreia var. argyreia Pan 1987, O. argyreia var. angustifolia has a linear-lanceolate to oblanceolate leaf blade and glabrous ovary while the leaf shape of O. argyreia var. argyreia is elliptic to ovate and comes with pubescent ovary (Wang et al. Citation1990). Taxonomic classification in Oreocharis remains challenging due to its species richness (Möller et al. Citation2016). Despite attempts at species delimitation of Oreocharis using morphological characteristics aided by geographical and molecular (i.e. DNA sequence data from short fragments) evidence, the relationship within Oreocharis and the molecular placement of O. argyreia var. angustifolia were only partly resolved at a nuclear level based on 573 nuclear orthologous genes (Kong et al. Citation2022; Yang et al. Citation2022); studies on the phylogenetic analysis of Oreocharis that used plastid gene sequence were in form of a combined dataset of trnL-trnF and the internal transcribed spacer (ITS) gene sequences, while the relationship in Oreocharis is still dubious at plastid level (Wang et al. Citation2010; Yang et al. Citation2020). We believe that the lack of informative sites in the DNA fragment dataset has hindered the understanding of this species at the genetic level. Thus, in this study, we sequenced and characterized the complete plastid genome (plastome) of O. argyreia var. angustifolia and reconstructed a phylogenetic tree to reveal its molecular placement among related species.
Materials and methods
Fresh leaves of O. argyreia var. angustifolia were obtained from a natural population in Mount Shixia of Longnan town (24°55’37.79”, 114°51’5.46”). A voucher specimen (collection number: JLSXGL20220512) has been deposited in the Herbarium of Jiulian Mountain National Nature Reserve (contact: Guoliang Xu; email: [email protected]). As the species is not listed as a protected species, no permit is required for specimen collection.
Total genomic DNA was extracted using the cetyltrimethylammonium bromide (CTAB) method (Doyle and Doyle Citation1990). A 300-bp genomic library was constructed using the TruSeq DNA Sample Prep Kit (Illumina, USA), and sequencing was performed on an Illumina Novaseq platform. Plastome assembly was carried out on the generated 150-bp paired-end raw data using GetOrganelle v1.7.7.0 (Jin et al. Citation2020) based on default parameters. Gene annotation was performed using GeSeq v2.03 (Tillich et al. Citation2017) with the default parameters and was manually checked for errors. The annotated plastome and the structure of the genes that are difficult to annotate, including the cis-splicing and trans-splicing genes, were visualized using CPGView (http://www.1kmpg.cn/cpgview).
To infer the molecular placement of O. argyreia var. angustifolia among its relatives, phylogenetic reconstruction was carried out based on 79 unique CDS shared among the plastomes of 13 taxa of Gesneriaceae. Two species, Olea paniculata (Oleaceae; GenBank accession number: MT560031; Dong et al. Citation2022) and Plantago depressa (Plantaginaceae; GenBank accession number: MK144833; Kwon et al. Citation2019) were included as outgroups. The CDS were aligned using MAFFT 7.470 (Katoh and Standley Citation2013) before concatenation. Phylogenetic analysis was conducted using the maximum likelihood (ML) and Bayesian inference (BI) methods via the RAxML v8 (Stamatakis Citation2014) and MrBayes v3.2 (Ronquist et al. Citation2012) pipelines available in the CIPRES Science Gateway (Miller et al. Citation2010), respectively. The ML tree was constructed using the general-time reversible (GTR) with gamma distribution (+G) (=GTR + G) nucleotide substitution model, coupled with 1,000 bootstrap replicates. Markov chain Monte Carlo with 2,000,000 generations was used for the BI tree, and sampling was taken at every 100 cycles.
Results
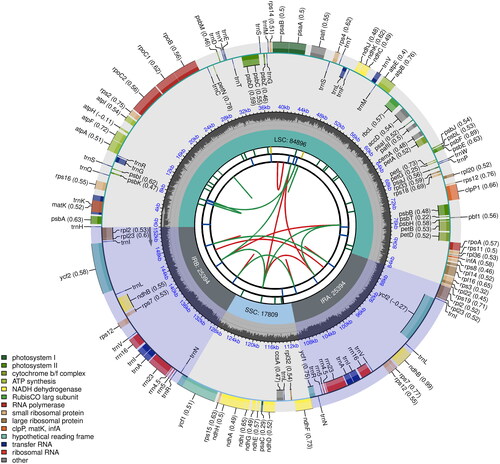
With the minimum read mapping depth of 30× and an average read mapping depth of 103.8× (Supplementary Figure 1), the complete plastome of O. argyreia var. angustifolia (GenBank accession number: ON872365) exhibits a typical quadripartite structure and was 153,493 bp in length (). The plastome includes a large single-copy (LSC) region of 84,896 bp, and a small single-copy (SSC) region of 17,809 bp, separated by a pair of 25,394-bp inverted repeat (IR) regions. A total of 131 genes were predicted, including 86 protein-coding, 37 tRNA, and eight rRNA genes. Among them, 13 CDS were cis-splicing genes, of which two contain two introns and 11 contain one intron (Supplementary Figure 2A). The gene structure of the trans-splicing gene, rps12 was also identified (Supplementary Figure 2B). The overall GC content was 37.6%.
Figure 2. Complete plastome map of Oreocharis argyreia var. angustifolia. Chloroplast genome map of Christella dentata. From the center outward, the first track shows the dispersed repeats, in which the forward (D) and palindromic (P) repeats are connected with red and green arcs. The second track shows the long tandem repeats as short blue bars. The third track shows the short tandem repeats or microsatellite sequences as short bars with different colors that correspond to their repeat unit size: Black: complex repeat; green: repeat unit size = 1; yellow: repeat unit size = 2; purple: repeat unit size = 3; blue: repeat unit size = 4; orange: repeat unit size = 5; red: repeat unit size = 6. The small single-copy, inverted repeat, and large single-copy regions are shown on the fourth track. The GC content along the genome is plotted on the fifth track. The genes are shown on the sixth track, while the optional codon usage bias is displayed in the parenthesis after the gene name. Genes are color-coded by their functional classification (bottom left corner), while the transcription directions for the inner and outer genes are clockwise and anticlockwise, respectively.
As both the ML and BI trees showed the same topology, only the ML tree was displayed; the posterior probabilities of the BI analysis were incorporated into the ML tree (). The phylogenetic relationship among genera in Gesneriaceae was well-resolved (bootstrap support, BS ≥75%; posterior probability, PP ≥0.95). Based on current circumscription, the Oreocharis clade is placed sister to Hemiboea ovalifolia. Among the four Oreocharis species, O. chienii was the first to diverge, while followed by O. argyreia var. angustifolia and O. esquirolii, as well as the other two species, O. cotinifolia and O. mileensis.
Figure 3. Phylogenetic tree based on the 79 shared unique CDS of 13 selected taxa of Gesneriaceae, with Olea paniculata and Plantago depressa included as outgroups. Shown next to the nodes are the bootstrap support (BS) and posterior probability (PP) values, in which strong branch support (BS ≥75%, left; PP ≥0.95, right) is indicated with an asterisk (*).

Discussion
When compared with the available plastomes of three other Oreocharis species, all four displayed a similar total number of genes and genome structure (Meng et al. Citation2019; Gu et al. Citation2020; Tang et al. Citation2021). The phylogenetic position of the four species of Orecharis included in this study was incongruent when compared to the phylogenetic tree reconstructed using the 573 nuclear orthologous genes (Yang et al. Citation2022). Based on the nuclear tree, two distinct clades were identified, in which O. argyreia var. angustifolia is clustered with O. continifolia and O. esquirolii, while O. mileensis was placed in another clade; species of Oreocharis is monophyletic when using the complete plastome sequences. Cyto-nuclear discordance is expected in Oreocharis as there is evidence of hybridization at the species level (Puglisi et al. Citation2011). As demonstrated in this study, a resolved tree was reconstructed for Gesneriaceae using the complete plastome sequence; thus, it shows the potential of the plastome tree to provide greater resolution when compared to the short gene sequences, i.e., trnL-trnF and ITS. Therefore, it is wise to consider the use of the complete plastome sequence to perform phylogenetic reconstruction of Orecharis as well as Gesneriaceae, suggesting a considerably enlarged sampling size to uncover the maternal relationship among members of Gesneriaceae.
Authors’ contributions
GX, SYL, XK: conception and design; GX, ZL, ZC, SYL: analysis and interpretation of data; GX, ZL, ZC: drafting of the paper; SYL, XK: critical revision of the paper; all the authors approved the final version; and all authors agree to be accountable for all aspects of the work.
Supplemental Material
Download TIFF Image (2.3 MB)Supplemental Material
Download TIFF Image (379.6 KB)Disclosure statement
No potential conflict of interest was reported by the author(s). Oreocharis argyreia var. angustifolia is not a protected plant. The study did not incur any disturbance or damages to its population; thus, no specific permissions are required.
Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI at http://www.ncbi.nlm.nih.gov under the accession number ON872365. The associated BioProject, SRA, and BioSample numbers are PRJNA772562, SRR19924869, and SAMN29409930, respectively.
Additional information
Funding
References
- Dong WP, Sun JH, Liu YL, Xu C, Wang YH, Suo ZL, Zhou SL, Zhang ZX, Wen J. 2022. Phylogenomic relationships and species identification of the olive genus Olea (Oleaceae). J of Syt Evol. 60(6):1263–1280. doi: 10.1111/jse.12802.
- Doyle JJ, Doyle JL. 1990. Isolation of plant DNA from fresh tissue. Focus. 12:39–40.
- Fan Q, Zhao WY, Shi S, Jing HJ, Huang ZF, Liao WB. 2014. Additions to the seed plant flora of Jiangxi Province. China. Subtropical Plant Sci. 43(1):29–32.
- Gu L, Su T, An MT, Hu GX. 2020. The complete chloroplast genome of the vulnerable Oreocharis esquirolii (Gesneriaceae): structural features, comparative and phylogenetic analysis. Plants. 9(12):1692. doi: 10.3390/plants9121692.
- Jin JJ, Yu WB, Yang JB, Song Y, DePamphilis CW, Yi TS, Li DZ. 2020. GetOrganelle: a fast and versatile toolkit for accurate de novo assembly of organelle genomes. Genome Biol. 21(1):241. doi: 10.1186/s13059-020-02154-5.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780. doi: 10.1093/molbev/mst010.
- Kong H, Condamine FL, Yang L, Harris AJ, Feng C, Wen F, Kang M. 2022. Phylogenomic and macroevolutionary evidence for an explosive radiation of a plant genus in the Miocene. Syst Biol. 71(3):589–609. doi: 10.1093/sysbio/syab068.
- Kwon W, Kim Y, Park CH, Park J. 2019. The complete chloroplast genome sequence of traditional medical herb, Plantago depressa Willd. (Plantaginaceae). Mitochondrial DNA Part B: resour. 4(1):437–438. doi: 10.1080/23802359.2018.1553530.
- Li S, Xin ZB, Su LY. 2018. The importance of protection and conservation for Gesneriaceae from the endangered categories of new published taxa in China. China’s Bot. Gard. 21:24–35.
- Meng J, Zhang LN, He J. 2019. Complete plastid genome of the endangered species Paraisometrum mileense (Gesneriaceae) endemic to China. Mitochondrial DNA Part B Resour. 4(2):3585–3586. doi: 10.1080/23802359.2019.1677186.
- Miller MA, Pfeiffer W, Schwartz T. 2010. Creating the CIPRES science gateway for inference of large phylogenetic trees. Proceedings of the Gateway Computing Environments Workshop (GCE), 14 November 2010, IEEE, New Orleans.
- Möller M, Wei YG, Wen F, Clark JL, Weber A. 2016. You win some you lose some: updated generic delineations and classification of Gesneriaceae-implications for the family in China. Guihaia. 36(1):44–60.
- Mou C, Peng CL, Zhang F. 2019. Analysis of species diversity of Gesneriaceae plants in Hunan. Guangxi Sci. 26(1):141–145.
- POWO. Plants of the World Online. 2022. Facilitated by the Royal Botanic Gardens, Kew. http://www.plantsoftheworldonline.org.
- Puglisi C, Wei YI-G, Nishii K, Möller M. 2011. Oreocharis× heterandra (Gesneriaceae): a natural hybrid from the Shengtangshan Mountains, Guangxi, China. Phytotaxa. 38(1):1–18. doi: 10.11646/phytotaxa.38.1.1.
- Ronquist F, Teslenko M, Van Der Mark P, Ayres DL, Darling A, Höhna S, Larget B, Liu L, Suchard MA, Huelsenbeck JP. 2012. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol. 61(3):539–542. doi: 10.1093/sysbio/sys029.
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30(9):1312–1313. doi: 10.1093/bioinformatics/btu033.
- Tang JL, Zhao B, Li CL, Hong X. 2021. Complete chloroplast genome sequence of an endangered plant Oreocharis cotinifolia (Gesneriaceae) from Guangxi, China. Mitochondrial DNA Part B Resour. 6(10):2936–2938. doi: 10.1080/23802359.2021.1973918.
- Tillich M, Lehwark P, Pellizzer T, Ulbricht-Jones ES, Fischer A, Bock R, Greiner S. 2017. GeSeq – versatile and accurate annotation of organelle genomes. Nucleic Acids Res. 45(W1):W6–W11. doi: 10.1093/nar/gkx391.
- Wang WC, Pan KY, Li ZY, Weitzman AL, Skog LE. 1990. Gesneriaceae. In: Wang WC. (eds.), Flora of China, vol. 18, p. 251–261. Science Press, Beijing & Missouri Botanical Garden, St. Louis.
- Wang YZ, Liang RH, Wang BH, Li JM, Qiu ZJ, Li ZY, Weber A. 2010. Origin and phylogenetic relationships of the Old World Gesneriaceae with actinomorphic flowers inferred from ITS and trnL-trnF sequences. Taxon. 59(4):1044–1052. doi: 10.1002/tax.594005.
- Xu WB, Guo J, Pan B, Zhang Q, Liu Y. 2017. Diversity and distribution of Gesneriaceae in China. Guihaia. 37(10):1219–1226.
- Yang J-W, Qin X-M, Xu J, Li C-R, Ren Q-F, Yuan M-Q, Zhang Q, Yi S-R, Cai L. 2022. Oreocharis qianyuensis, a new species of Gesneriaceae from Southwest, China based on morphological and molecular evidence. PhytoKeys. 213:119–130. doi: 10.3897/phytokeys.213.84349.
- Yang LH, Wen F, Kong HH, Sun ZX, Su LY, Kang M. 2020. Two new combinations in Oreocharis (Gesneriaceae) based on morphological, molecular and cytological evidence. PhytoKeys. 157:43–58. doi: 10.3897/phytokeys.157.32609.

