Abstract
A recently published complete mitochondrial genome of Japanese or Temminck’s cormorant (Phalacrocorax capillatus) was the first of this species (GenBank accession number LC714913). Comparison of COI sequences shows that this mitogenome clustered with great cormorant (Phalacrocorax carbo) rather than with its sister taxon P. capillatus. This suggests that the mitogenome was either a misidentified P. carbo or represents previously unknown intraspecific diversity in P. capillatus overlapping with that of P. carbo. Unfortunately, no voucher specimen was retained so it remains impossible to distinguish between these alternatives. We suggest that great restraint should be exercised using this mitogenome as a reference for P. capillatus. We reiterate previous pleas to retain voucher specimens for mitogenome sequences to enable re-analysis of the identity of the material.
Introduction
Japanese or Temminck’s Cormorant (Phalacrocorax capillatus) (Temminck & Schlegel, 1850) is a coastal waterbird breeding along northeast Asian coasts, from southeast Russia including Sakhalin, south Kuril Islands, Hokkaido to Kyushu, Japan, the Korean Peninsula and northeast China. In most parts of its range, including Japan, it co-exists with Great Cormorant P. carbo (Lethaby and Moores Citation1999; Squires et al. Citation2022). The two species are closely related (Kennedy and Spencer Citation2014; Kennedy et al. Citation2023). P. capillatus and P. carbo are very similar in morphology but can be distinguished by subtle differences in shape, structure, and the coloration of the plumage and bare parts (Lethaby and Moores Citation1999).
The first complete mitochondrial genome sequence (hereafter mitogenome) of P. capillatus was published by Honda et al. (Citation2022). This sequence was derived from a blood sample taken from an individual rescued in Aomori City, northern Honshu, Japan, which died a few days later in captivity (GenBank accession number LC714913). Honda et al. (Citation2022) included a phylogram based on complete mitogenomes which, as expected, placed LC714913 sister to a mitogenome of P. carbo (KR215630). However, the two sequences showed almost zero divergence suggesting that the two sequences may belong to the same species.
Materials and methods
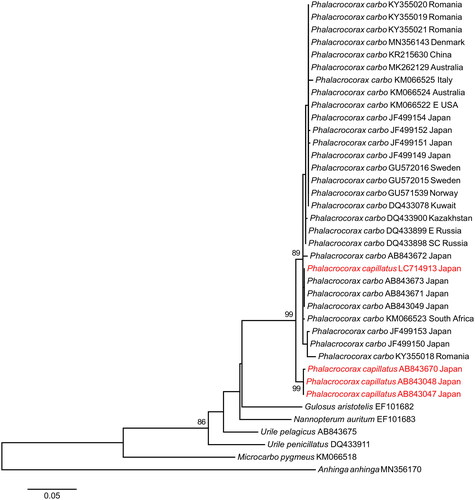
We verified the identity of LC714913 by comparing 696 bp of the cytochrome c oxidase subunit I of the mitogenome to reference sequences of P. capillatus (n = 3) and P. carbo (n = 28, including several from Japan). A maximum-likelihood phylogeny was obtained using MEGA7 with 1000 bootstrap replicates (Kumar et al. Citation2016). Model testing was performed in MEGA7. The appropriate substitution model, GTR + G + I, was selected based on the Akaike information criterion. Sequence divergence was calculated as uncorrected p values with complete deletion of nucleotide positions with missing data.
Results
In the COI phylogeny (), P. carbo and P. capillatus formed well-supported reciprocally monophyletic groups. However, LC714913 was placed within the P. carbo clade with strong support (bootstrap support 89%). Mean sequence divergence of P. carbo and P. capillatus was 1.3%. In both species, mean intraspecific sequence divergence was 0.1%. Mean divergence between LC714913 and P. carbo was 0.2%, whereas mean divergence between LC714913 and P. capillatus was 1.4%.
Discussion
Based on current knowledge of sequence variation in P. carbo and P. capillatus, the phylogenetic position of LC714913 among P. carbo and its low sequence divergence with P. carbo (similar to mean intraspecific sequence divergence of P. carbo) suggest that LC714913 is a misidentified P. carbo rather than P. capillatus.
Sequence variation in P. capillatus is known from only three bona fide sequences from a single study (Saitoh et al. Citation2015). Thus, it is possible that sequence variation in P. capillatus is greater than presently known and perhaps overlaps with that of P. carbo. If this is the case, LC714913 could be a valid sequence of P. capillatus. It is known that some valid species of birds are reciprocally non-monophyletic in mitochondrial sequences (e.g. Joseph et al. Citation2009; Kerr et al. Citation2009; Techow et al. Citation2010).
A voucher specimen is necessary to distinguish between these two possibilities. Unfortunately, no voucher specimen of LC714913 was preserved (Honda et al. Citation2022). Without a voucher specimen, the specific identity of LC714913 may long remain uncertain, diminishing its value for population genetic, conservation genetic, and phylogenetic studies. The species identity of the sample used to generate LC714913 may be inferred from nuclear DNA genotyping, but this requires nuclear DNA sequences of P. capillatus that are not currently available. Unless and until the species identity of this sample is ascertained, we suggest that great restraint should be exercised using mitogenome LC714913 as a reference for P. capillatus.
Given that most of the recently published mitogenomes are intended to serve as the reference sequence for population genetic, conservation genetic, and phylogenetic studies, it is important that the identity of mitogenomes is established beyond doubt and can be verified after the publication of the sequence. Recent studies have unveiled many misidentified or otherwise problematic mitogenomes, showing that the identity of published mitogenomes cannot be taken at face value (Botero-Castro et al. Citation2016; Oleinik et al. Citation2019; Sangster and Luksenburg Citation2020, Citation2021a, Citation2021b, Citation2021c). The present case shows that DNA identification is sometimes insufficient to establish the identity of a sequence. In such cases, it is necessary to revisit the original specimen. The importance of voucher specimens has been pointed out before in systematics (Peterson et al. Citation2007; Pleijel et al. Citation2008), DNA barcoding (Collins and Cruickshank Citation2013) and mitogenomics (Strohm et al. Citation2016). Irrespective of the availability of a voucher specimen, we argue that all mitogenome announcements should include a statement about how the sample was identified, including any diagnostic character states of the relevant animal. As pointed out by Sangster and Luksenburg (Citation2021a), such information can easily be included in the main text or in the online supporting materials. If no specimen could be preserved, photographs of the sampled individual should be included illustrating the diagnostic character states for the relevant taxon.
A third possibility is that the similarity of mitogenome LC714913 to P. carbo rather than to P. capillatus is due to introgression from P. carbo into P. capillatus. In this scenario, the phenotype of the bird would have been similar or identical to P. capillatus but the nuclear genome would be mixture of both species. Analysis of nuclear DNA would be able to verify this scenario but, again, this requires sequences that are not currently available for P. capillatus.
Author contributions
George Sangster: conceptualization, methodology, formal analysis, investigation, and writing – original draft. Jolanda A. Luksenburg: writing – review and editing. Both authors agreed to be accountable for all aspects of the work and approved the final draft to be published.
Acknowledgements
We thank Atsushi Sogabe and two anonymous referees for their constructive comments that helped improve the paper.
Disclosure statement
No potential conflict of interest was reported by the authors.
Data availability statement
The sequence data that support the findings of this study were published previously and are openly available on GenBank at https://www.ncbi.nlm.nih.gov/nucleotide. All sequences used in this study are referenced in and can be retrieved by typing in the relevant GenBank accession number(s) in the search panel on the aforementioned webpage. The mitochondrial genome of Phalacrocorax capillatus (LC714913) is available at https://www.ncbi.nlm.nih.gov/nuccore/LC714913.
Additional information
Funding
References
- Botero-Castro F, Delsuc F, Douzery EJ. 2016. Thrice better than once: quality control guidelines to validate new mitogenomes. Mitochondrial DNA A DNA Mapp Seq Anal. 27(1):449–454. doi: 10.3109/19401736.2014.900666.
- Collins RA, Cruickshank RH. 2013. The seven deadly sins of DNA barcoding. Mol Ecol Res. 13:969–975.
- Honda R, Inumaru M, Sato Y, Sogabe A. 2022. Complete mitochondrial genome of the Japanese Cormorant Phalacrocorax capillatus (Temminck & Schlegel, 1850) (Suliformes: Phalacrocoracidae). Mitochondrial DNA B Resour. 7(8):1577–1578. doi: 10.1080/23802359.2022.2113753.
- Joseph L, Adcock GJ, Linde C, Omland KE, Heinsohn R, Chesser RT, Roshier D. 2009. A tangled tale of two teal: population history of the grey Anas gracilis and chestnut teal A. castanea of Australia. J Avian Biol. 40(4):430–439. doi: 10.1111/j.1600-048X.2008.04652.x.
- Kennedy M, Salis AT, Seneviratne SS, Rathnayake D, Nupen LJ, Ryan PG, Volponi S, Lubbe P, Rawlence NJ, Spencer HG. 2023. Phylogeny of the microcormorants, with the description of a new genus. Zool J Linn Soc. 199(1):310–317. doi: 10.1093/zoolinnean/zlad041.
- Kennedy M, Spencer HG. 2014. Classification of the cormorants of the world. Mol Phylogenet Evol. 79:249–257. doi: 10.1016/j.ympev.2014.06.020.
- Kerr KC, Birks SM, Kalyakin MV, Red’kin YA, Koblik EA, Hebert PD. 2009. Filling the gap – CO1 barcode resolution in eastern Palearctic birds. Front Zool. 6(1):29. doi: 10.1186/1742-9994-6-29.
- Kumar S, Stecher G, Tamura K. 2016. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 33(7):1870–1874. doi: 10.1093/molbev/msw054.
- Lethaby N, Moores N. 1999. Identification of Temminck’s cormorant. Dutch Bird. 21(1):1–8.
- Oleinik AG, Skurikhina LA, Kukhlevsky AD. 2019. Clarification of taxonomic assignment of smelt complete mitochondrial genome: GenBank accession number KP281293.1 (NC_026566.1). Mitochondrial DNA Part B. 4(1):1696–1697. doi: 10.1080/23802359.2019.1607578.
- Peterson AT, Moyle RG, Nyári AS, Robbins MB, Brumfield RT, Remsen JV. 2007. The need for proper vouchering in phylogenetic studies of birds. Mol Phylogenet Evol. 45(3):1042–1044. doi: 10.1016/j.ympev.2007.08.019.
- Pleijel F, Jondelius U, Norlinder E, Nygren A, Oxelman B, Schander C, Sundberg P, Thollesson M. 2008. Phylogenies without roots? A plea for the use of vouchers in molecular phylogenetic studies. Mol Phylogenet Evol. 48(1):369–371. doi: 10.1016/j.ympev.2008.03.024.
- Saitoh T, Sugita N, Someya S, Iwami Y, Kobayashi S, Kamigaichi H, Higuchi A, Asai S, Yamamoto Y, Nishiumi I. 2015. DNA barcoding reveals 24 distinct lineages as cryptic bird species candidates in and around the Japanese Archipelago. Mol Ecol Resour. 15(1):177–186. doi: 10.1111/1755-0998.12282.
- Sangster G, Luksenburg JA. 2020. The published complete mitochondrial genome of Eptesicus serotinus is a chimera of Vespertilio sinensis and Hypsugo alaschanicus (Mammalia: Chiroptera). Mitochondrial DNA B Resour. 5(3):2661–2664. doi: 10.1080/23802359.2020.1785349.
- Sangster G, Luksenburg JA. 2021a. Sharp increase of problematic mitogenomes of birds: causes, consequences, and remedies. Genome Biol Evol. 13(9):evab210.
- Sangster G, Luksenburg JA. 2021b. The published complete mitochondrial genome of milk shark (Rhizoprionodon acutus) is a misidentified Pacific spadenose shark (Scoliodon macrorhynchos) (Chondrichthyes: Carcharhiniformes). Mitochondrial DNA B Resour. 6(3):828–830. doi: 10.1080/23802359.2021.1884019.
- Sangster G, Luksenburg JA. 2021c. Scientific data laundering: chimeric mitogenomes of a sparrowhawk and a nightjar covered-up by forged phylogenies. Biochem Syst Ecol. 96:104263. doi: 10.1016/j.bse.2021.104263.
- Squires TE, Aoki D, Hasegawa O. 2022. The recent range expansion of great cormorants Phalacrocorax carbo in Hokkaido, Japan. Ardea. 109(3):389–394. doi: 10.5253/arde.v109i2.a11.
- Strohm JH, Gwiazdowski RA, Hanner R. 2016. Mitogenome metadata: current trends and proposed standards. Mitochondrial DNA A DNA Mapp Seq Anal. 27(5):3263–3269. doi: 10.3109/19401736.2015.1015003.
- Techow NMSM, O'Ryan C, Phillips RA, Gales R, Marin M, Patterson-Fraser D, Quintana F, Ritz MS, Thompson DR, Wanless RM, et al. 2010. Speciation and phylogeography of giant petrels Macronectes. Mol Phylogenet Evol. 54(2):472–487. doi: 10.1016/j.ympev.2009.09.005.