Abstract
Semiothisa cinerearia Bremer & Grey, 1853 belongs to the lepidopteran family Geometridae, subfamily Ennominae. We sequenced the complete mitochondrial genome of S. cinerearia by PCR and Sanger sequencing method. The mitochondrial genome of S. cinerearia is 15,523 bp in length and contains a typical set of 37 genes with ‘MIQ’ type gene arrangement and a 394 bp AT-rich regions. Except for cox1 using CGA as initiation codon, all protein-coding genes (PCGs) start with ATN codons and except for nad2 and nad4l using TAG as termination codon, all PCGs terminated with TAA codon. A phylogenetic tree including 39 genus of subfamily Ennominae was first reconstructed based on the mitochondrial genome sequences with nucleotide substitution model GTR + G + I, which showed that the genera Amraica, Jankowskia, and Ectropis are not monophyletic and S. cinerearia and Macaria notata are classified together.
1. Introduction
The moth of Semiothisa cinerearia Bremer & Grey, 1853 has gray and yellow brown body, filiform antennas and about 30–45 mm wingspan. Both the subbasal and the medial lines in the forewings are dark brown and turn into acute corners at the part near outer margins. The subterminal lines on the forewings are black-brown, consist of two or three rows of black-brown plaques and form a single brown triangular patch at the part near outer margins. A rectangular brown patch is out of the subterminal line. The apex of the forewings is pale yellowish brown with a dark triangular patch below it (). The moths distribute widely in Japan and China (Beijing, Hebei, Shandong, Jiangsu, Zhejiang, Jiangxi, Taiwan, Shaanxi, Gansu, and Xizang). The larvae of S. cinerearia are harmful mainly to landscape plant Chinese scholar tree Sophora japonica L., 1767 and sometimes to Robinia pseudoacacia L., 1753. Lepidopteran mitochondrial genomes, containing conservative 37 genes, which are important for constructing electron transport chains and ATP synthases, have been wildly used for the phylogenetic analysis at the family or subfamily level. About one hundred of mitochondrial genomes of subfamily Ennominae have been sequenced and 13 articles had been published (Yang et al. Citation2013; Liu et al. Citation2014; Xu et al. Citation2016; Sun et al. Citation2017; Wang et al. Citation2017; Li et al. 2018; Chen et al. Citation2019; Du et al. Citation2019; Liu et al. Citation2020; Huang et al. Citation2021; Song et al. Citation2021; Sun et al. Citation2021; Chen et al. Citation2022). While subfamily Ennominae contains about 1100 genera and 9700 described species, more mitochondrial genomes should be sequenced for comprehensive understanding of the phylogeny of the subfamily. Here, we report the complete mitochondrial genome of S. cinerearia of subfamily Ennominae, and try to perform a phylogenetic analysis based on the known mitochondrial genomes to clarify the branch relationships among these Ennominae species.
2. Materials and methods
2.1. Sample collection, DNA extraction, and preservation
The specimen of S. cinerearia was light-trapped from Xiangshan Mountain, Huaibei City, Anhui Province, China (latitude 116.816834, longitude 33.989836). The total DNA was extracted from the muscle of the specimen legs according to the instruction manual of Ezup Column Animal Genomic DNA Purification Kit (Sangon, Shanghai, China) and the quantity and quality of the extracted DNA were evaluated by NanoDrop 2000c spectrophotometer (Thermo, Waltham, MA) and 1% agarose gel electrophoresis. The specimen (accession number 20170722E) and the DNA solution (accession number DNA20170722E) were deposited in the Specimens Room and the Human and Animal Genetics Laboratory (contact person LiJun and email [email protected]), College of Life Sciences, Huaibei Normal University, China.
2.2. Mitochondrial DNA amplification, sequencing, assemblage, and annotation
Primers to amplify mitochondrial DNA were designed according to the conserve regions of published lepidopteran mitochondrial genomes and produced by Sangon Biotech (Shanghai) Co., Ltd. (Shanghai, China). The overlapping fragments were amplified using PrimeSTAR GXL DNA Polymerase (Takara, Beijing, China) and sequenced with Sanger dideoxy sequencing method by Shanghai Sequencing Department of Beijing Genomics Institution (BGI) (Shanghai, China). Complete mitochondrial DNA sequence was assembled using Lasergene DNASTAR7.1 (DNAStar, Madison, WI) (Burland Citation2000), preliminarily annotated using MITOS2 (Bernt et al. Citation2013) and manually verified with NCBI BLAST.
2.3. Sequence alignment and phylogenic tree reconstitution
Ninety-nine mitochondrial genomes of subfamily Ennominae have been published in GenBank up to now, and most of which are neither annotated nor published in unified arrangement. These mitochondrial genomes were downloaded and only one mitochondrial genome per species was retained and rearranged into unified sequence with the first nucleotide of tRNA-Met as the start position after aligned with reference sequence such as NC_069306. Sequences were aligned using MAFFT7.505 with L-INS-I method (Katoh et al. Citation2002). The phylogenetic tree was reconstructed using Bayesian inference (BI) method with software MrBayes3.2.6 (Ronquist and Huelsenbeck Citation2003; Ronquist et al. Citation2012). Iotaphora admirabilis Oberthür, 1883 (Geometrinae) and Idaea salutaria Christoph, 1881 (Sterrhinae) were used as outgroups (Ding et al. Citation2020). The best nucleotide substitution model was assessed by Mega11.0 with the lowest Bayesian information criterion (BIC) score (Tamura et al. Citation2021).
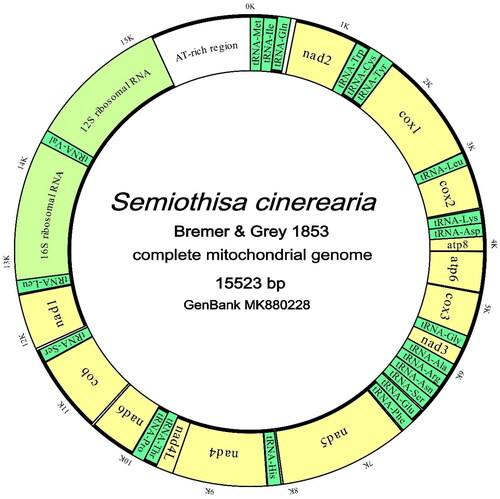
Figure 2. Complete mitochondrial genome map of S. cinerearia with 13 protein coding genes, 22 tRNAs, 2 rRNAs, and an AT-rich region. Outside and inner circles represent the J- and M-strand respectively. Bold lines in circles represent strands in which the genes lie. (Green: tRNAs; yellow: PCGs; greenish yellow: rRNAs; grey: overlaps; white: interval spaces).
3. Results and discussion
3.1. Characteristics of mitochondrial genome
The complete mitochondrial genome of S. cinerearia (GenBank accession number MK880228) is 15,523 bp, and contains 13 protein-coding genes (PCGs), 22 transfer RNA genes (tRNAs), two ribosomal RNA genes (rRNAs), and a 394 bp AT-rich regions. Twenty-three genes are located on the majority strand (J-strand), in which nine are PCGs and 14 are tRNAs. The remaining 14 genes are located on the minority strand (N-strand), in which four are PCGs, eight are tRNAs and two are rRNAs. The mitochondrial genome characteristics usually refer to the characteristics of its J-strand. The nucleotide composition of the J-strand of the mitochondrial genome is A 41.50%, C 12.09%, G 7.94%, and T 38.47%. The gene arrangement is just as most of lepidopteran mitochondrial genomes with ‘MIQ’ arrangement and 17 intergenic spacers (228 bp in total) and eight overlaps (59 bp in total) were founded (). The intergenic nucleotides vary from 1 to 59 bp and the longest intervals are located between tRNA-Gln and nad2, while the overlap nucleotides vary from 1 to 20 bp and the maximum overlap lies between cox2 and tRNA-Lys (). Except for cox1 using CGA as initiation codon, all PCGs start with ATN codons. Except for nad2 and nad4l using TAG as termination codon, all PCGs terminated with TAA codon. The AT-rich region contains the classic ‘ATAGA + polyT’ motif, two ‘TA’ tandem repeats and a polyA tail.
Table 1. Annotations of the mitochondrial genome of S. cinerearia (GenBank MK880228).
3.2. Phylogenetic analysis
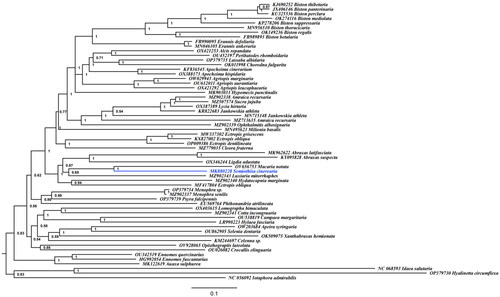
The mitochondrial genomes for reconstructing the phylogenetic tree of subfamily Ennominae belong to 39 genera, 58 species, and the best nucleotide substitution model is GTR + G + I. The tree shows that neither genus Amraica nor Jankowskia was classified together. Ectropis obliqua (MF417804) was not classified into the clade of genus Ectropis, which was consistent with the result obtained by Sun et al. (Citation2021). Semiothisa cinerearia and Macaria notata were classified together ().
Figure 3. The Bayesian inference (BI) phylogenetic tree was reconstructed based on published mitochondrial genome sequences of subfamily Ennominae and two species of subfamily Geometrinae were used as outgroup. GenBank accession numbers were indicated before the species names. Numbers at the nodes indicated Bayesian posterior probability values.
4. Conclusions
The gene arrangement of the mitochondrial genome of S. cinerearia is ‘M-I-Q’ model and S. cinerearia is closer to M. notata than other 57 known mitochondrial genomes of Ennominae species. The genera Amraica, Jankowskia, and Ectropis are not monophyletic, which indicates the classification within subfamily Ennominae should be further studied and more mitochondrial genome data are needed.
Ethics statement
The experiments were approved by the Animal Ethics Committee of Huaibei Normal University and conducted following the Chinese Association for the Laboratory Animal Sciences and the Institutional Animal Care and Use Committee (IACUC) protocols.
Author contributions
Deng Zou, Jun Yuan, and Jie Ding involved in the sample collection, DNA extraction, and PCR amplification. Jun Li and Haijun Zhang involved in primer design, data analysis, and draft writing. All authors have read and agreed to the final version of the manuscript and to be accountable for all aspects of the work.
Disclosure statement
No potential conflict of interest was reported by the authors.
Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI at https://www.ncbi.nlm.nih.gov/ under the accession no. MK880228. The associated BioProject and Bio-Sample numbers are PRJNA946324 and SAMN33819875, respectively. This mitochondrial genome was not obtained by NGS and does not show the SRA number here.
Additional information
Funding
References
- Bernt M, Donath A, Jühling F, Externbrink F, Florentz C, Fritzsch G, Pütz J, Middendorf M, Stadler PF. 2013. MITOS: improved de novo metazoan mitochondrial genome annotation. Mol Phylogenet Evol. 69(2):313–319. doi: 10.1016/j.ympev.2012.08.023.
- Burland TG. 2000. DNASTAR's Lasergene sequence analysis software. Bioinform Methods Protoc. 132:71–91.
- Chen HY, Li W, Shen S. 2022. Characterization and phylogenetic analysis of the mitochondrial genome of Xanthabraxas hemionata (Lepidoptera: Geometridae). Mitochondrial DNA B Resour. 7(8):1481–1483. doi: 10.1080/23802359.2022.2107447.
- Chen YM, Wang QH, Wang SB, Qu LJ. 2019. The mitochondrial genome of Erannis ankeraria (Lepidoptera: Geometridae). Mitochondrial DNA B Resour. 4(2):2696–2697. doi: 10.1080/23802359.2019.1644560.
- Ding JH, Yang Y, Li J. 2020. Complete mitochondrial genome of Iotaphora admirabilis (Lepidoptera: Geometridae). Mitochondrial DNA B Resour. 5(2):1425–1426. doi: 10.1080/23802359.2020.1736958.
- Du YM, Song X, Lu ZJ. 2019. Phylogenetic relationship and characterization of the complete mitochondrial genome of Milionia basalis (Lepidoptera: Geometridae). Mitochondrial DNA B Resour. 4(2):4136–4137. doi: 10.1080/23802359.2019.1692732.
- Huang BS, Zhao YJ, Wu YP. 2021. Complete mitochondrial genome of Biston thoracicaria (Lepidoptera: Geometridae). Mitochondrial DNA B Resour. 6(7):2007–2008. doi: 10.1080/23802359.2021.1939181.
- Katoh K, Misawa K, Kuma K, Miyata T. 2002. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 30(14):3059–3066. doi: 10.1093/nar/gkf436.
- Li Q, Wang X, Chen X, Han B. 2018. Complete mitochondrial genome of the tea looper caterpillar, Ectropis obliqua (Lepidoptera: Geometridae) with a phylogenetic analysis of Geometridae. Int J Biol Macromol. 114:491–496. doi: 10.1016/j.ijbiomac.2018.02.038.
- Liu HL, Chen ZT, Liu C, Wu XL, Xiao KJ, Mao JH, Pu DQ. 2020. Mitochondrial genomic analysis of the tea geometrid, Ectropis grisescens Warren, 1894 (Lepidoptera: Geometridae) and its phylogenetic implications. J Entomol Res Soc. 22:325–337.
- Liu SX, Xue DY, Cheng R, Han HX. 2014. The complete mitogenome of Apocheima cinerarius (Lepidoptera: Geometridae: Ennominae) and comparison with that of other lepidopteran insects. Gene. 547(1):136–144. doi: 10.1016/j.gene.2014.06.044.
- Ronquist F, Huelsenbeck JP. 2003. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 19(12):1572–1574. doi: 10.1093/bioinformatics/btg180.
- Ronquist F, Teslenko M, van der Mark P, Ayres DL, Darling A, Hohna S, Larget B, Liu L, Suchard MA, Huelsenbeck JP. 2012. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol. 61(3):539–542. doi: 10.1093/sysbio/sys029.
- Song XH, Yang TB, Xu XQ, Yan XH, Zhou CQ. 2021. Characterization of the complete mitochondrial genome of Ectropis grisescens (Lepidoptera, Geometridae). Mitochondrial DNA B Resour. 6(7):1953–1955. doi: 10.1080/23802359.2021.1923423.
- Sun Y, Wang J, Li F, Huang X, Geng X, Li J. 2021. Complete mitochondrial genome of Hypomecis punctinalis Scopoli, 1763 and its phylogenetic position within family Geometridae. Mitochondrial DNA B Resour. 6(10):2910–2912. doi: 10.1080/23802359.2021.1969698.
- Sun Y, Zhang JW, Li QQ, Liang D, Abbas MN, Qian C, Wang L, Wei GQ, Zhu BJ, Liu CL. 2017. Mitochondrial genome of Abraxas suspecta (Lepidoptera: Geometridae) and comparative analysis with other Lepidopterans. Zootaxa. 4254(5):501–519. doi: 10.11646/zootaxa.4254.5.1.
- Tamura K, Stecher G, Kumar S. 2021. MEGA11 Molecular Evolutionary Genetics Analysis Version 11. Mol Biol Evol. 38(7):3022–3027. doi: 10.1093/molbev/msab120.
- Wang XQ, Chen SC, Li PW, Hu X, Peng P. 2017. The complete mitochondrial genome of a tea geometrid, Ectropis obliqua (Lepidoptera: Geometridae). Mitochondrial DNA B Resour. 2(2):459–460. doi: 10.1080/23802359.2017.1357449.
- Xu YM, Chen SC, Wang XQ, Peng P, Li PW. 2016. The complete mitogenome of Jankowskia athleta (Lepidoptera: Geometridae). Mitochondrial DNA A DNA Mapp Seq Anal. 27(4):3035–3036. doi: 10.3109/19401736.2015.1063122.
- Yang XS, Xue DY, Han HX. 2013. The complete mitochondrial genome of Biston panterinaria (Lepidoptera: Geometridae), with phylogenetic utility of mitochondrial genome in the Lepidoptera. Gene. 515(2):349–358. doi: 10.1016/j.gene.2012.11.031.

