Abstract
This study determined the complete mitochondrial genome of the jellyfish Pelagia noctiluca (Scyphozoa, Semaeostomeae) for the first time. The genome was a linear molecule of 16,390 bp in length and 59.3% AT. It comprised of 13 typical protein-coding genes (cox1-3, nd1-6, nd4L, atp6, atp8, and cytB), two ribosomal RNAs (16S and 12S rRNA), and two tRNAs (trnM and trnW). In addition, we detected two additional open reading frames (polB and ORF314) at one end of the genome. The gene-coding structures were identical to those of other scyphozoans. Based on a molecular phylogeny constructed using 13 protein-coding genes, P. noctiluca has the closest genetic relationship with the genus Chrysaora (Semaeostomeae).
Introduction
The jellyfish Pelagia noctiluca Forskål, 1775 (Cnidaria, Scyphozoa) is usually pink or mauve in color and has a phosphorescent bell, tentacles, and oral arms (). It is globally distributed in warm and temperate waters (Kramp Citation1961; Russell Citation1970), and blooms of the organism have been constantly recorded (Canepa et al. Citation2014; Ramesh et al. Citation2022). It has negative impact on fisheries, mariculture, and tourism (Purcell et al. Citation1999, Citation2007). In addition, the jellyfish can sting swimmers, causing local symptoms such as pain, erythema, edema, and/or vesicles (Cegolon et al. Citation2013). Their sting incidents and blooms are reported as some of the most serious in the Mediterranean Sea (Brotz and Pauly Citation2012; Canepa et al. Citation2014).
Figure 1. Image of an adult Pelagia noctiluca. It shows mauve in color and has a phosphorescent bell with lappets and tentacles at the edge and oral arms. The photograph was taken in 2008, by Hans Hillewaert (adopted from wikipedia).

Pelagia noctiluca belongs to the family Pelagiidae, and it is the only described species within the genus Pelagia. Phylogenetic relationships of the Pelagia and relatives have been inferred using mitochondrial (mt) genes, including cytochrome c oxidase subunit I (cox1) and 16S ribosomal DNA (Bayha et al. Citation2017). In recent years, whole mt genomes of Pelagiidae jellyfishes have been used for constructing deeper phylogenetic relationships (Curole and Kocher Citation1999; Feng et al. Citation2023). Previously, Kayal et al. determined a partial mt genome of P. noctiluca (15,876 bp; GenBank accession number: JN700949) and used it to study evolution in medusozoan cnidarians (Kayal et al. Citation2012). However, the genome was a partial sequence, and thus it has not been completely elucidated, especially at both ends due to the linear structure of the genome. In the present study, we determined the complete mt genome sequence of Korean P. noctiluca and characterized the protein-coding genes (PCGs), particularly considering two extra open reading frames (polB and ORF314) and the complete cytB.
Materials and methods
Specimens of Pelagia noctiluca were collected from the southern sea (34°21'11.7" N, 127°31'00.8" E) in South Korea on May 26, 2021. The collected samples were fixed with 100% ethanol. The sample and gDNA were stored in the specimen room of the Department of Biotechnology, Sangmyung University, Korea (Dr. Han-Sol Kim, [email protected]) under the voucher number FM113.
Genomic DNA was extracted with the cetyl trimethylammonium bromide (CTAB) method (Richards et al. Citation2003). Based on the available partial mt genome sequence and structure of P. noctiluca (JN700949), we designed two primers that targeted the terminus genes polB and cytB and two primers that targeted nd2 (). Two primer pairs (JF-F1 × JF-R1 and JF-F2 × JF-R2) were used to amplify the nearly complete mt genome with a long and accurate polymerase chain reaction (LA PCR). The PCR products were confirmed with electrophoresis on a 1.5% agarose gel (Figure S1). The sequences were obtained with PCR primers and primer walking (Table S1) through Sanger sequencing via the ABI3730 DNA sequencer (Applied Biosystems, Foster City, CA). In addition, the terminus of the genome was determined with the single-primed PCR method (Loh Citation1991) using terminal deoxynucleotidyl transferase (Takara Shuzo Co., Kyoto, Japan), cloned using TOPcloner TA kit (Enzynomics Inc., Daejeon, Korea), and subjected to sequencing (). All the sequences of the mt genome were assembled using Sequencher 5.1 (Gene Codes Corporation, Ann Arbor, MI) with the following assembly parameters: Minimum Match Percentage: 90%, and Minimum Overlap: 16 (Figure S2). The full-length sequence of the mt genome was annotated using MITOS (Bernt et al. Citation2013) and Geneious 9.1.3 (Geneious, Auckland, New Zealand).
Table 1. Primer sets for PCR; each set was for the middle and ends of the genome.
We generated a molecular phylogenetic tree for the class Scyphozoa using 12 complete mt genomes, including the outgroup hydrozoan Craspedacusta sowerbii. It was constructed by using the concatenated amino acid sequences of 13 PCGs of the mt genome. The phylogenetic analysis was conducted with the maximum-likelihood (ML) method with 1,000 bootstrap replicates based on LG + G + I + F model using MEGA7 (Kumar et al. Citation2016).
Results
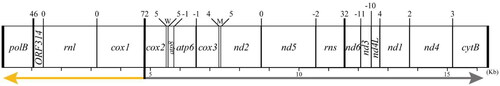
The total length of the mt genome of Korean Pelagia noctiluca (GenBank accession number: OQ446325) was determined to be 16,390 bp (26.5% A, 32.7% T, 19.9% C, and 20.7% G). A linearly formed genome consisted of 13 typical PCGs (cox1-3, nd1-6, nd4L, atp6, atp8, and cytB), two ribosomal RNAs (16S and 12S rRNA, rnl and rns), two transfer RNAs (tRNA) (trnM and trnW), and two extra open reading frames (ORFs), polB and ORF314 (). The gene order of the mt genome in P. noctiluca was identical to that of other scyphozoans. There were two start codons ATG and GTG (cox3 and nd1) and two stop codons TAG (polB, ORF314, cox1, atp6, and nd4L) and TAA in the genome. In addition, five overlapping regions between atp8 and atp6 (1 bp), atp6 and cox3 (1 bp), nd5 and 12S rRNA (2 bp), nd6 and nd3 (11 bp), and nd3 and nd4L (10 bp) were detected.
Figure 2. Gene map of the mitochondrial genome of the species. It forms a linear structure and is composed of 15 protein-coding genes, 2 tRNAs (M and W), and 2 rRNAs. Upper numbers indicate intergenic spacers, and lower numbers and graduations indicate the length. Lower arrows provide the orientation of the strands (yellow: reverse, gray: forward).
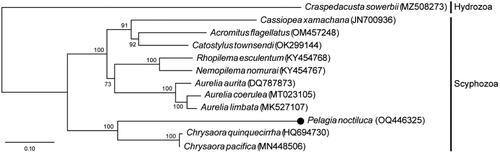
The phylogenetic tree indicates two major clades within scyphozoans (). One clade consisted of six genera (Cassiopea, Acromitus, Catostylus, Nemopilema, Rhopilema, and Aurelia), and the other clustered with two Pelagiidae members, Chrysaora and Pelagia.
Figure 3. A maximum-likelihood (ML) tree of the class Scyphozoa was constructed using the 13 concatenated mitochondrial protein-coding genes from the complete mt genomes and LG + G + I + F model. We used the following sequences: Craspedacusta sowerbii MZ508273 (unpublished), Cassiopea xamachana JN700936 (unpublished), Acromitus flagellatus OM457248 (Lin et al. Citation2022), Catostylus townsendi OK299144 (unpublished), Rhopilema esculentum KY454768 (Wang and Sun Citation2017b), Nemopilema nomurai KY454767 (Wang and Sun Citation2017a), Aurelia aurita DQ787873 (Shao et al. Citation2006), A. coerulea MT023105 (Seo et al. Citation2020), A. limbate MK527107 (Karagozlu et al. Citation2019), Chrysaora quinquecirrha HQ694730 (Park et al. Citation2012), and C. pacifica MN448506 (Wang and Yin Citation2020). Pelagia noctiluca in the present study is marked with a black dot.
Discussion and conclusion
The mt genomes of medusozoan cnidarians (e.g. cubozoans, staurozoans, and hydrozoans) have class-specific elements such as linear structure and two additional ORFs (Bridge et al. Citation1992, Citation1995; Kayal et al. Citation2012). The two ORFs polB and ORF314 were predicted to have respective functions as B-type DNA polymerase and terminal protein, possibly attributing to protecting each terminus in the jellyfish’s mt genome (Kayal et al. Citation2012). Because of the linear structure of the genome and the terminal location of the extra genes, they have not been thoroughly investigated in P. noctiluca. This is the first study to determine the full-length sequences of the polB (993 bp, 56.8% AT) and cytB (1,137 bp, 59.9% AT) genes in P. noctiluca. In addition, compared to the previous study, we newly determined the stop codons of ORF314, nd5, and cytB (TAA) and polB (TAG) and the overlapping region between nd5 and 12S rRNA (2 bp). All genes were investigated completely in the P. noctiluca mt genome. However, we did not find telomere regions or something similar at the ends of the mt genome, of which patterns were detected in some linear genomes of jellyfishes (Smith et al. Citation2012).
In conclusion, the linear mt genome sequence of Korean P. noctiluca was determined, and the gene sequence was characterized for the first time. The structure and gene order of the genome were common with other scyphozoans (Kayal et al. Citation2012). In the ML phylogenetic analysis, P. noctiluca showed the closest relationship with Chrysaora, consistent with a previous study (Kayal et al. Citation2015). The present result will provide important information for phylogeographic and genetic studies of Pelagiidae.
Ethical approval
The mauve stinger jellyfish Pelagia noctiluca Forskål, is not an Endangered or protected species, instead harmful organism; therefore, specific permission was not required to collect this species. Research on this species, including the collection of specimens, was conducted following the guidelines provided by Sangmyung University and Korean Government.
Author contributions
HE Lee: Experiments, data analyses, and original draft preparation. JS Ki: Conceptualization, Supervision, conceived and designed project, and writing-reviewing and editing.
Supplemental Material
Download MS Word (2.9 MB)Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The data supporting this study’s findings are openly available in GenBank with the accession number OQ446325. The associated BioProject, BioSample, and SRA numbers are PRJNA1024046, SAMN37687440, and SRR26283628 to SRR26283655, respectively.
Additional information
Funding
References
- Bayha KM, Collins AG, Gaffney PM. 2017. Multigene phylogeny of the scyphozoan jellyfish family Pelagiidae reveals that the common US Atlantic Sea nettle comprises two distinct species (Chrysaora quinquecirrha and C. chesapeakei). PeerJ. 5:e3863. doi: 10.7717/peerj.3863.
- Bernt M, Donath A, Jühling F, Externbrink F, Florentz C, Fritzsch G, Pütz J, Middendorf M, Stadler PF. 2013. MITOS: improved de novo metazoan mitochondrial genome annotation. Mol Phylogenet Evol. 69(2):313–319. doi: 10.1016/j.ympev.2012.08.023.
- Bridge D, Cunningham CW, Schierwater B, Desalle ROB, Buss LW. 1992. Class-level relationships in the phylum Cnidaria: evidence from mitochondrial genome structure. Proc Natl Acad Sci USA. 89(18):8750–8753. doi: 10.1073/pnas.89.18.8750.
- Bridge D, Cunningham CW, DeSalle R, Buss LW. 1995. Class-level relationships in the phylum Cnidaria: molecular and morphological evidence. Mol Biol Evol. 12(4):679–689.
- Brotz L, Pauly D. 2012. Jellyfish populations in the Mediterranean Sea. Acta Adriat. 53(2):213–232.
- Canepa A, Fuentes V, Sabatés A, Piraino S, Boero F, Gili JM. 2014. Pelagia noctiluca in the Mediterranean Sea. In: Pitt KA ,Lucas CH, editors. Jellyfish blooms. Dordrecht (The Netherlands): Springer Science + Business Media; p. 237–266.
- Cegolon L, Heymann WC, Lange JH, Mastrangelo G. 2013. Jellyfish stings and their management: a review. Mar Drugs. 11(2):523–550. doi: 10.3390/md11020523.
- Curole JP, Kocher TD. 1999. Mitogenomics: digging deeper with complete mitochondrial genomes. Trends Ecol Evol. 14(10):394–398. doi: 10.1016/s0169-5347(99)01660-2.
- Feng H, Lv S, Li R, Shi J, Wang J, Cao P. 2023. Mitochondrial genome comparison reveals the evolution of cnidarians. Ecol Evol. 13(6):e10157. doi: 10.1002/ece3.10157.
- Karagozlu MZ, Seo Y, Ki JS, Kim CB. 2019. The complete mitogenome of brownbranded moon jellyfish Aurelia limbata (Cnidaria, Semaeostomeae, Ulmaridae) with phylogenetic analysis. Mitochondrial DNA B: resour. 4(1):1875–1876. doi: 10.1080/23802359.2019.1614494.
- Kayal E, Bentlage B, Collins AG, Kayal M, Pirro S, Lavrov DV. 2012. Evolution of linear mitochondrial genomes in medusozoan cnidarians. Genome Biol Evol. 4(1):1–12. doi: 10.1093/gbe/evr123.
- Kayal E, Bentlage B, Cartwright P, Yanagihara AA, Lindsay DJ, Hopcroft RR, Collins AG. 2015. Phylogenetic analysis of higher-level relationships within Hydroidolina (Cnidaria: hydrozoa) using mitochondrial genome data and insight into their mitochondrial transcription. PeerJ. 3:e1403. doi: 10.7717/peerj.1403.
- Kramp PL. 1961. Synopsis of the medusae of the world. J Mar Biol Ass. 40:7–382. doi: 10.1017/S0025315400007347.
- Kumar S, Stecher G, Tamura K. 2016. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 33(7):1870–1874. doi: 10.1093/molbev/msw054.
- Lin J, Feng S, Wang L, Qiu Y. 2022. Complete mitochondrial genome sequence of Acromitus flagellatus and its phylogenetic relationship with related jellyfish species. Mitochondrial DNA B Resour. 7(10):1823–1824. doi: 10.1080/23802359.2022.2131367.
- Loh E. 1991. Anchored PCR: amplification with single-sided specificity. Methods. 2(1):11–19. doi: 10.1016/S1046-2023(05)80121-5.
- Park E, Hwang DS, Lee JS, Song JI, Seo TK, Won YJ. 2012. Estimation of divergence times in cnidarian evolution based on mitochondrial protein-coding genes and the fossil record. Mol Phylogenet Evol. 62(1):329–345. doi: 10.1016/j.ympev.2011.10.008.
- Purcell JE, Malej A, Benović A. 1999. Potential links of jellyfish to eutrophication and fisheries. Ecosystems at the Land‐Sea Margin: drainage Basin to Coastal Sea. 55:241–263.
- Purcell JE, Uye SI, Lo WT. 2007. Anthropogenic causes of jellyfish blooms and their direct consequences for humans: a review. Mar Ecol Prog Ser. 350:153–174. doi: 10.3354/meps07093.
- Ramesh CH, Koushik S, Shunmugaraj T, Ramana Murthy MV. 2022. Occurrence of a scyphozoan jellyfish, Pelagia noctiluca (Forskål, 1775) bloom in the Gulf of Mannar Marine National Park, Southern India. Indian J Mar Sci. 50(2):161–164.
- Richards E, Reichardt M, Rogers S. 2003. Preparation of genomic DNA from plant tissue. In: Ausubel FM, Brent R, Kingston RE, Moore DD, Seidman JG, Smith JA, Struhl K, editors. Current protocols in molecular biology. New York: John Wiley and Sons; p. 2.3.1–2.3.7.
- Russell FS. 1970. The Medusae of the British Isles volume II: pelagic scyphozoa, with a supplement to the first volume of Hydromedusae. London: Cambridge University Press; p. 1–284.
- Seo Y, Chae J, Ki JS. 2020. The complete mitochondrial genome of the jellyfish Aurelia coerulea (Cnidaria and Scyphozoa) with phylogenetic analysis. Mitochondrial DNA B: resour. 5(2):1929–1930. doi: 10.1080/23802359.2020.1749155.
- Smith DR, Kayal E, Yanagihara AA, Collins AG, Pirro S, Keeling PJ. 2012. First complete mitochondrial genome sequence from a box jellyfish reveals a highly fragmented linear architecture and insights into telomere evolution. Genome Biol Evol. 4(1):52–58. doi: 10.1093/gbe/evr127.
- Shao Z, Graf S, Chaga OY, Lavrov DV. 2006. Mitochondrial genome of the moon jelly Aurelia aurita (Cnidaria, Scyphozoa): a linear DNA molecule encoding a putative DNA-dependent DNA polymerase. Gene. 381:92–101. doi: 10.1016/j.gene.2006.06.021.
- Wang Y, Sun S. 2017a. Complete mitochondrial genome of the jellyfish, Nemopilema nomurai (Cnidaria: Scyphozoa) and the phylogenetic relationship in the related species. Mitochondrial DNA B Resour. 2(1):165–166. doi: 10.1080/23802359.2017.1298417.
- Wang Y, Sun S. 2017b. Complete mitochondrial genome of the jellyfish, Rhopilema esculentum Kishinouye 1891 (Cnidaria: Scyphozoa) and the phylogenetic relationship in the related species. Mitochondrial DNA B Resour. 2(1):167–168. doi: 10.1080/23802359.2017.1303342.
- Wang Y, Yin J. 2020. Complete mitochondrial genome of the jellyfish, Chrysaora pacifica (Goette, 1886) (Cnidaria, Scyphozoa) and the phylogenetic relationship in the related species. Mitochondrial DNA B Resour. 5(1):455–456. doi: 10.1080/23802359.2019.1704648.
