Abstract
Acanthogobius lactipes is a demersal, euryhaline fish belonging to the suborder Gobiodei. This study sequenced and described the complete mitochondrial genome of A. lactipes for the first time. The circular genome of A. lactipes is 16,592 bp in length and contains 13 protein-coding genes, 22 transfer RNA genes, two ribosomal RNA genes, and a control region. The overall A, C, G, and T contents were 27.78, 27.31, 17.52, and 27.39%, respectively. Based on the 13 protein-coding genes, the phylogenetic tree showed that A. lactipes formed a well-supported cluster with the genus Acanthogobius and rooted with other family Oxudercidae species.
1. Introduction
The white-limbed goby, Acanthogobius lactipes (Hilgendorf, 1879) is a demersal, euryhaline fish widely distributed in the Japan, Korean Peninsula, China, and Russia, and lives mainly in estuaries (Hosoya Citation2015; Chae et al. Citation2019). This species belongs to the suborder Gobioidei (Gobiiformes), a group containing over 2,000 species worldwide, and belongs to the family Oxudercidae within this suborder (Nelson et al. Citation2016). Recently, large-scale phylogenetic studies of the suborder Gobioidei using molecular tools provided evidence that it is reasonable to divide the suborder Gobioidei into two major clades: the Gobiidae and Oxudercidae (Thacker Citation2003, Citation2009, Citation2013; Agorreta et al. Citation2013; Nelson et al. Citation2016). However, the Gobiidae and Oxudercidae family members remain controversial because they are diverse with many subgroups, and a few lineages remain unresolved (Akihito et al. Citation2000; McCraney et al. Citation2020). Mitochondrial DNA is a reliable molecular marker for studying phylogenetic relationships because it has the unique characteristics of maternal inheritance, a fast rate of evolution, and a high copy number (Avise et al. Citation1987; Boore et al. Citation2005). We described the complete mitochondrial genome sequence and structure of A. lactipes for the first time by next-generation sequencing (NGS) and investigated the phylogenetic position of this species within the Gobiidae and Oxudercidae family members. This study will contribute to our understanding of the phylogenetic relationship and evolutionary history of A. lactipes belonging to the suborder Gobioidei and provide a genetic source for further research.
2. Materials and methods
2.1. Sample collection and preservation
A specimen of A. lactipes () was collected with skimming nets from the downstream basin of the Daecheon Stream located in Boryeong-si, Chungcheongnam-do, Republic of Korea, on July 4, 2022 (36°21′7.05″N, 126°33′35.71″E). The specimen was preserved in 99% ethanol and deposited at the specimen storage facility of Soonchunhyang University (Prof. I.-C. Bang, [email protected]) under voucher number SUC26331.
Figure 1. The specimen of Acanthogobius lactipes from the Daecheon Stream, Boryeong-si, Chungcheongnam-do, Republic of Korea (photo by Bong Han Yun). the main identifiable morphological characteristics are the dorsal fin VIII-IX, I 9-11; anal fin I 9-11; scales in the longitudinal row 34-38; the predorsal scales 0-9; and several irregular dark blotches along the mid-body.

2.2. DNA extraction and sequencing
Genomic DNA (gDNA) isolation was performed using the phenol-chloroform extraction procedure (Taggart et al. Citation1992). A genomic library for the NGS was prepared from the isolated gDNA using the MGIEasy DNA Library Prep Kit (MGI Tech Co. Ltd., Shenzhen, China). NGS raw data were obtained by paired-end reads (2 × 150 bp) on an MGISEQ-2000 sequencing platform, trimmed with Cutadapt 4.2 (Martin Citation2011), and a contig sequence was assembled using CLC Genomics Workbench 20.04 (CLC Inc., Aarhus, Denmark). The circular form of the mitochondrial genome was confirmed by mapping the clean data onto a contig sequence using Geneious R11 (Kearse et al. Citation2012) (Figure S1). We annotated the final sequence using the MITOS web-based tool (Bernt et al. Citation2013). Finally, the complete genome sequence of the circular form was deposited in the GenBank database (acc. no. OR238485). The genome map was generated using the OGDRAW program (Greiner et al. Citation2019).
2.3. Phylogenetic analysis
We collected 13 protein-coding gene (PCGs) sequences from the mitochondrial genome of species belonging to the Gobiidae and Oxudercidae families from GenBank (Table S1) to determine the phylogenetic position of A. lactipes. Two species belonging to the family Butidae were used as the outgroup, and the collected sequences were aligned using MAFFT 7.475 (Katoh and Standley Citation2013). GTR + G + I was selected as the optimal substitution model based on the corrected Akaike information criterion (AICc) estimated by jModelTest 2.1.10 (Darriba et al. Citation2012). A Bayesian inference (BI) tree was reconstructed using Mrbayes 3.2.7 (Ronquist et al. Citation2012), run for 2,000,000 generations, and sampled every 1,000 steps with 25% of the initial trees discarded as burn-in.
3. Results
3.1. Mitogenomic characterization
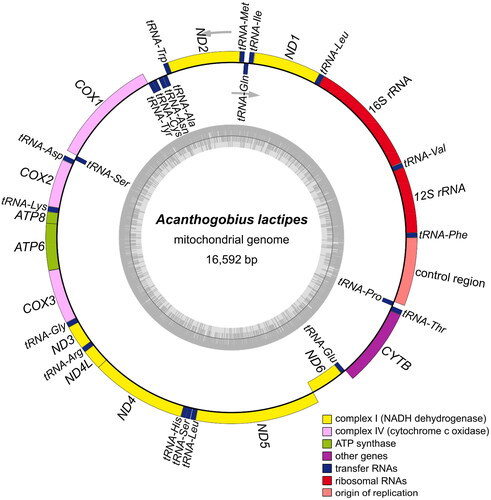
The complete mitochondrial genome of A. lactipes was 16,592 bp (OR238485). The mitogenome comprised 13 PCGs, 22 transfer RNA (tRNA) genes, two ribosomal RNA (rRNA) genes, and one control region (). The overall A, C, G, and T contents were 27.78, 27.31, 17.52, and 27.39%, respectively, with a slight A + T bias of 55.17%.
The total PCGs of A. lactipes was 11,369 bp, accounting for 68.81% of the complete mitogenome and encoding 3,796 amino acids. Of the 13 PCGs, all were encoded on the H-strand except for ND6, which was encoded on the L-strand. Except for the COX1 gene starting with a GTG start codon, the remaining 12 PCGs started with an ATG start codon. Seven PCGs terminated with complete stop codons (TAA or TAG), while the COX2, COX3, CYTB, ND2, ND3, and ND4 genes terminated with incomplete stop codons (T or TA). The lengths of the 12S and 16S rRNA genes were 952 and 1,673 bp, respectively. The control region was located between tRNAPro and tRNAPhe and was 991 bp. The 22 tRNA genes varied in length from 66 to 75 bp.
3.2. Phylogenetic analysis
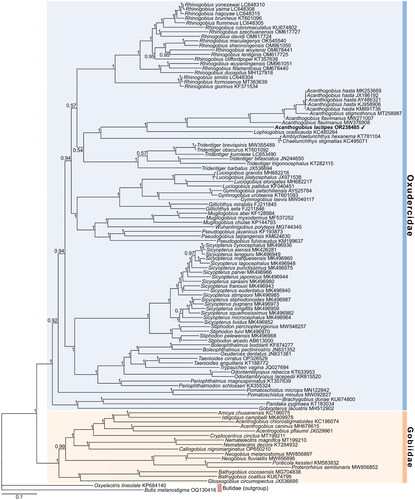
The constructed phylogenetic tree showed a marked phylogenetic divergence between the Gobiidae and Oxudercidae families with high statistical support (Bayesian posterior probabilities (BPP) = 1). It showed A. lactipes as a member of the Oxudercidae (). Furthermore, A. lactipes clustered with other Acanthogobius species, showing that this cluster was closely related to Lophiogobius ocellicauda, Amblychaeturichthys hexanema, and Chaeturichthys stigmatias (BPP = 1).
4. Discussion and conclusion
This study is the first to identify the complete mitochondrial genome of A. lactipes using NGS technology. The gene order and content of the A. lactipes mitogenome were identified as being like other Gobioidei mitogenomes published previously (Kim et al. Citation2004; Wang et al. Citation2019; Nam and Rhee Citation2020). As described previously (Thacker Citation2003; Agorreta et al. Citation2013), the phylogenetic tree showed a large group, which until recently, was classified as the suborder Gobioidei and was divided into two main clades: the Gobiidae and Oxudercidae. In addition, the results showing A. lactipes clustering with other fish of the genus Acanthogobius and being closely related to L. ocellicauda, A. hexanema, and C. stigmatias were similar to the phylogenetic analysis by McCraney et al. (Citation2020) using some nuclear and mitochondrial DNA genes. A. lactipes was reportedly morphologically similar to Acanthogobius luridus inhabiting the Korean Peninsula and China (Lee Citation1992; Chae et al. Citation2019). Although they are similar in body shape and coloration, A. luridus is distinguished by cycloid scales on the top of the opercular and occipital regions (Lee Citation1992). However, the complete mitochondrial genome of A. luridus has yet to be explored. Therefore, to understand A. lactipes and related species fully, the mitogenome of A. luridus must be identified, and molecular phylogenetic analysis must be performed. In conclusion, the complete mitochondrial genome provided in this study is a solid basis for further studies of the genome-based phylogenetic and evolutionary relationships in A. lactipes and related species.
Author contributions statement
Bong Han Yun, Yong Hwi Kim, and In-Chul Bang conceived the original idea. Bong Han Yun carried out the experiments. Bong Han Yun wrote the manuscript with support from Yong Hwi Kim, Jong Yeon Park, Duce Tam Huynh, and In-Chul Bang. Jong Yeon Park, Ho-Seop Han, Hye Jin Kim, and Ho Sung Lee analyzed the experimental data. All authors have agreed to be accountable for all aspects of the work.
Ethical approval
The sample used in this study was a dead body of fish, and as per the animal experimental ethics of the Republic of Korea (standard operating guideline; IACUC - Institutional Animal Care and Use Committee, Book no. 11-1543061-000457-01, effective Dec. 2020) does not need any approval from an ethics committee. Additionally, the species used in this study is not endangered fish, and the sampling site is not located in any protected area.
Supplemental Material
Download MS Word (1.4 MB)Disclosure statement
No potential conflict of interest was reported by the authors.
Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI at [https://www.ncbi.nlm.nih.gov] (https://www.ncbi.nlm.nih.gov/) under the accession no. OR238485. The associated BioProject, SRA, and Bio-Sample numbers are PRJNA1007762, SRR25713998, and SAMN37097069 respectively.
Additional information
Funding
References
- Agorreta A, San Mauro D, Schliewen U, Van Tassell JL, Kovačić M, Zardoya R, Rüber L. 2013. Molecular phylogenetics of Gobioidei and phylogenetic placement of European gobies. Mol Phylogenet Evol. 69(3):619–633. doi:10.1016/j.ympev.2013.07.017.
- Akihito, Iwata A, Kobayashi T, Ikeo K, Imanishi T, Ono H, Umehara Y, Hamamatsu C, Sugiyama K, Ikeda Y, Sakamoto K, Fumihito A, Ohno S, Gojobori T. 2000. Evolutionary aspects of gobioid fishes based upon a phylogenetic analysis of mitochondrial cytochrome b genes. Gene. 259(1–2):5–15. doi:10.1016/s0378-1119(00)00488-1.
- Avise JC, Arnold J, Ball RM, Bermingham E, Lamb T, Neigel JE, Reeb CA, Saunders NC. 1987. Intraspecific phylogeography: the mitochondrial DNA bridge between population genetics and systematics. Annu Rev Ecol Syst. 18(1):489–522. doi:10.1146/annurev.es.18.110187.002421.
- Bernt M, Donath A, Jühling F, Externbrink F, Florentz C, Fritzsch G, Pütz J, Middendorf M, Stadler PF. 2013. MITOS: improved de novo metazoan mitochondrial genome annotation. Mol Phylogenet Evol. 69(2):313–319. doi:10.1016/j.ympev.2012.08.023.
- Boore JL, Macey JR, Medina M. 2005. Sequencing and comparing whole mitochondrial genomes of animals. Methods Enzymol. 395:311–348.
- Chae BS, Song HB, Park JY, Cho KH, Kim IS, Cho SJ. 2019. A field guide to the freshwater fishes of Korea. Seoul, Korea: LG Evergreen Foundation.
- Darriba D, Taboada GL, Doallo R, Posada D. 2012. jModelTest 2: more models, new heuristics and parallel computing. Nat Methods. 9(8):772–772. doi:10.1038/nmeth.2109.
- Greiner S, Lehwark P, Bock R. 2019. OrganellarGenomeDRAW (OGDRAW) version 1.3.1: expanded toolkit for the graphical visualization of organellar genomes. Nucleic Acids Res. 47(W1):W59–W64. doi:10.1093/nar/gkz238.
- Hosoya K. 2015. Freshwater fishes of Japan. Tokyo, Japan: yama-kei Publishers.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780. doi:10.1093/molbev/mst010.
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, et al. 2012. Geneious basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 28(12):1647–1649. doi:10.1093/bioinformatics/bts199.
- Kim IC, Kweon HS, Kim YJ, Kim CB, Gye MC, Lee WO, Lee YS, Lee JS. 2004. The complete mitochondrial genome of the javeline goby Acanthogobius hasta (Perciformes, Gobiidae) and phylogenetic considerations. Gene. 336(2):147–153. doi:10.1016/j.gene.2004.04.009.
- Lee YJ. 1992. A taxonomic study of the genera Acanthogobius and Synechogobius (Pisces: gobiidae) from Korea. Korean J Ichthyol. 4(2):1–25.
- Martin M. 2011. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J. 17(1):10–12. doi:10.14806/ej.17.1.200.
- McCraney WT, Thacker CE, Alfaro ME. 2020. Supermatrix phylogeny resolves goby lineages and reveals unstable root of Gobiaria. Mol Phylogenet Evol. 151:106862. doi:10.1016/j.ympev.2020.106862.
- Nam SE, Rhee JS. 2020. Complete mitochondrial genome of the fire goby, Nemateleotris magnifica (Perciformes, Gobiidae). Mitochondrial DNA B Resour. 5(2):1894–1896. doi:10.1080/23802359.2020.1751738.
- Nelson JS, Grande T, Wilson MVH. 2016. Fishes of the world. 5th ed. Hoboken (NJ): John Wiley and Sons.
- Ronquist F, Teslenko M, van der Mark P, Ayres DL, Darling A, Höhna S, Larget B, Liu L, Suchard MA, Huelsenbeck JP. 2012. MrBayes 3.2: efficient bayesian phylogenetic inference and model choice across a large model space. Syst Biol. 61(3):539–542. doi:10.1093/sysbio/sys029.
- Taggart JB, Hynes RA, Prodöuhl PA, Ferguson A. 1992. A simplified protocol for routine total DNA isolation from salmonid fishes. J Fish Biol. 40(6):963–965. doi:10.1111/j.1095-8649.1992.tb02641.x.
- Thacker CE. 2003. Molecular phylogeny of the gobioid fishes (Teleostei: perciformes: gobioidei). Mol Phylogenet Evol. 26(3):354–368. doi:10.1016/s1055-7903(02)00361-5.
- Thacker CE. 2009. Phylogeny of Gobioidei and placement within Acanthomorpha, with a new classification and investigation of diversification and character evolution. Copeia. 2009(1):93–104. doi:10.1643/CI-08-004.
- Thacker CE. 2013. Phylogenetic placement of the European sand gobies in Gobionellidae and characterization of gobionellid lineages (Gobiiformes: gobioidei). Zootaxa. 3619(3):369–382. doi:10.11646/zootaxa.3619.3.6.
- Wang D, Dai C, Li Q, Li Y, Liu Z. 2019. Complete mitochondrial genome and phylogenic analysis of Rhinogobius cliffordpopei (Perciformes, Gobiidae). Mitochondrial DNA B Resour. 4(2):2473–2474. doi:10.1080/23802359.2019.1637287.