Abstract
The species of Saussurea sagittifolia Y. S. Chen & S. R. Yi belongs to the family Asteraceae (Cardueae). The complete chloroplast genome of S. sagittifolia was assembled and annotated for the first time in this study. The complete chloroplast genome of S. sagittifolia was 152,535 bp, including a large single-copy (LSC) region of 83,511 bp, a small single-copy (SSC) region of 18,632 bp, and a pair of inverted repeats (IRs) of 25,196 bp. The overall GC content of the chloroplast genome was 37.7%. The chloroplast genome encoded 131 genes, including 87 protein-coding genes, 36 tRNA genes, and eight rRNA genes. Phylogenetic analysis based on complete chloroplast sequences revealed that it related closely to Saussurea medusa.
Introduction
The Saussurea is one of the largest genera in the tribe Cardueae of family Asteraceae, with highly medicinal and ornamental value in China (Shi and Raab-Straube Citation2011; Li et al. Citation2022). It is mainly distributed in Asia, Europe, and North America (Shi and Raab-Straube Citation2011). Saussurea sagittifolia Y. S. Chen & S. R. Yi is a new species from the Bashan Mountains region in North Sichuan province, China in 2020 (Xu et al. Citation2020). The species of S. sagittifolia is close to S. oligocephala in morphological characteristics. However, it is different with the basal leaves triangular-ovate, a margin of small sharp teeth, apex acuminate, base with an auricular protrusion, and cauline leaves of more than 10, triangular, lanceolate, linear to subulate (Xu et al. Citation2020). In this study, we collected this species in Guangwu Shan Mountain, Bazhong City, Sichuan Province, China. We sequenced and reported the complete chloroplast genome of S. sagittifolia using next-generation sequencing technology for the first time. It will be helpful for well understanding the species delimitation studies and phylogenetic position of the genus Saussurea and S. sagittifolia in the Asteraceae.
Materials and methods
Sampling, extraction, and genome sequencing
The fresh leaves and specimen of S. sagittifolia () were collected from Guangwu Shan in Nanjiang County, Sichuan Province, China (106°48′3.16″E, 32°39′45.8″N). The voucher specimen was deposited at the Herbarium of Sichuan Normal University (SCNU) (Zhi-Xi Fu, [email protected]) under the voucher number: Ya Deng, DY158. Total genomic DNA was extracted using a modified CTAB method (Doyle and Doyle Citation1987). A paired-end library with an insert size of 150 bp was constructed, and the library was sequenced using Illumina NovaSeq 6000 platform at Beijing Genomics Institute (BGI, Shenzhen, China).
Figure 1. The morphology of leaf of S. sagittifolia. (a) The basal leaves of S. sagittifolia. (b) The leaves abaxially of S. sagittifolia. Leaf blades in middle and upper stem triangular, lanceolate, linear to subulate, apex long-caudate, base attenuate, margin with a few small sharp teeth to subentire. Leaves abaxially usually dark purple, capitula fewer, and involucres narrowly campanulate. The photos of S. sagittifolia were taken by the Zhi-Xi Fu in Guangwushan Mountain, Nanjiang county, Bazhong city, Sichuan province, China (the voucher number: Ya Deng, DY158).

Genome assembly and annotation
The high-quality reads were assembled with SPAdes v3.10.1 (Bankevich et al. Citation2012) and annotated by PGA (Qu et al. Citation2019) with default settings. To assess the accuracy of the assembly, we calculated the depth of coverage by mapping the reads to the chloroplast genome sequence using a specific protocol (https://doi.org/10.17504/protocols.io.4r3l27jkxg1y/v1). The annotation result was drawn using the CPGview program (http://www.1kmpg.cn/cpgview/) (Liu et al. Citation2023). The complete chloroplast sequence of S. sagittifolia deposited in GenBank of the National Center for Biotechnology Information (NCBI, https://www.ncbi.nlm.nih.gov, accession number ON094066).
Phylogenetic analysis
In order to reveal the phylogenetic position of S. sagittifolia with other members of Asteraceae, a phylogenetic analysis was performed based on 24 complete chloroplast genomes of Asteraceae, and Anthriscus cerefolium (Apiaceae), Kalopanax septemlobus (Araliaceae) as outgroups. Sequence alignment was achieved using the MAFFT (Katoh and Standley Citation2013). A maximum-likelihood (ML) analysis was carried out with RaxML v7.2.8 based on the GTRGAMMA model on the CIPRES (https://www.phylo.org/) using 1000 bootstrap replicates (Stamatakis Citation2014).
Results
Genome structure analysis
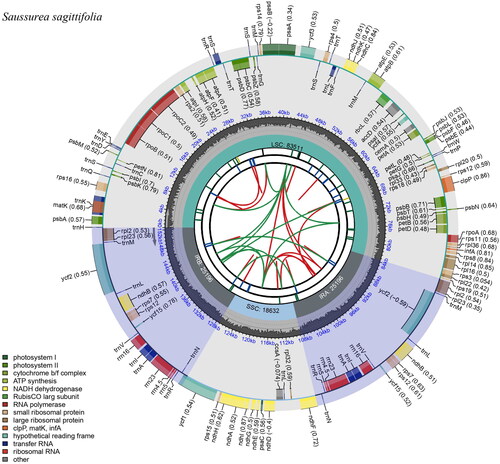
The clean reads of S. sagittifolia were approximately 5.7 Gb. The complete chloroplast genome of S. sagittifolia was 152,535 bp in length, with an average depth of ×223.80 (Supplementary Figure 1). It had a typical quadripartite structure, including a large single-copy region (LSC, 83,511 bp), a small single-copy region (SSC, 18,632 bp), and two inverted repeat regions (IRs, 25,196 bp). The genome contained 131 genes, including 87 protein-coding genes, 36 tRNA genes, and eight rRNA genes (). Most of the genes occurred in a single copy; however, seven protein-coding genes (ndhB, rpl2, rpl23, rps12, rps7, ycf15, and ycf2), eight tRNA genes (trnA-UGC, trnI, trnL-CAA, trnM-CAU, trnN-GUU, trnR-ACG, trnS-UGA, and trnV-GAC), and four rRNA genes (4.5S, 5S, 16S, and 23S) were duplicated. In total, 16 genes contained introns, while rps12, clpP, and ycf3 contained two introns. The rps12 gene was a trans-spliced gene, with its 5′ end in the LSC region and its 3′ end positioned within the two IR regions (Supplementary Figure 2). Gene structure analysis was done for rps16, rpoC1, atpF, ycf3, clpP, petB, petD, rpl16, rpl2, ndhB, and ndhA difficult to annotate genes (Supplementary Figure 3). The overall GC content was 37.7%, whereas the corresponding values of LSC, SSC, and IR regions were 35.79%, 31.34%, and 43.12%, respectively.
Figure 2. The chloroplast genome map of S. sagittifolia generated using CPGview. Boxes of different sizes and colors in the outermost circle represent genes and their lengths. Genes inside the circle are transcribed clockwise, and those on the outside are transcribed counter-clockwise. The grey area in the middle circle represents the variation of GC content at different positions, and the regions and lengths represented by the tetrameric structures (LSC, SSC, IRa, and IRb) are plotted in different colors on the inner circle.
Phylogenetic analysis
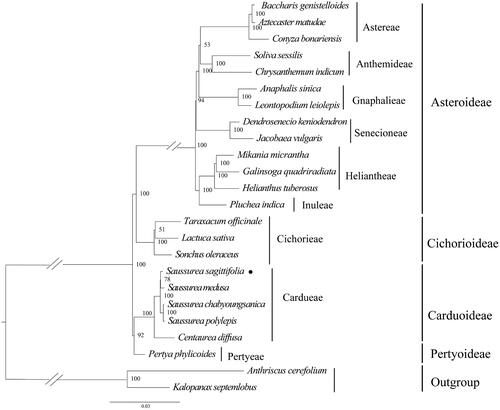
The phylogenetic analysis revealed that S. sagittifolia was belonging to the subfamily Carduoideae tribe Cardueae. Furthermore, S. sagittifolia was closely related to S. medusa. The genus Saussurea was recovered as a monophyletic group (). The chloroplast genome sequence of S. sagittifolia in this study might provide vital information for phylogenetic and evolutionary studies in Asteraceae.
Figure 3. Maximum-likelihood phylogeny of S. sagittifolia and related taxa based on 24 complete chloroplast genomes. Anthriscus cerefolium (Apiaceae), Kalopanax septemlobus (Araliaceae) were used as outgroups. The maximum-likelihood bootstrap support values are along the branches. Circles represent newly sequenced species. The following sequences were used: Aztecaster matudae KX063935 (Vargas et al. Citation2017), Baccharis genistelloides KX063864 (Vargas et al. Citation2017), Conyza bonariensis KX792499 (Wang et al. Citation2018), Chrysanthemum indicum JN867589 (unpublished), Soliva sessilis KX063863 (Vargas et al. Citation2017), Anaphalis sinica KX148081 (unpublished), Leontopodium leiolepis KM267636 (unpublished), Dendrosenecio keniodendron KY434193 (unpublished), Jacobaea vulgaris HQ234669 (Doorduin et al. Citation2011), Galinsoga quadriradiata KX752097 (Wang et al. Citation2018), Mikania micrantha KX154571 (Huang et al. Citation2016), Helianthus tuberosus MG696658 (unpublished), Pluchea indica MG452144 (Zhang et al. Citation2017), Taraxacum officinale KU361241 (unpublished), Lactuca sativa AP007232 (unpublished), Sonchus oleraceus MG878405 (Hereward et al. Citation2018), Saussurea sagittifolia ON094066 (in this study), Saussurea polylepis MF695711 (Seon et al. Citation2017), Centaurea diffusa KJ690264 (unpublished), Pertya phylicoides MN935435 (Wang et al. Citation2020), Anthriscus cerefolium GU456628 (Downie and Jansen Citation2015), and Kalopanax septemlobus NC022814 (Li et al. Citation2013).
Discussion and conclusions
In this study, the chloroplast genome sequence of S. sagittifolia was assembled and annotated for the first time. The genome size, organization, and structure were highly conserved and similar to other Asteraceae species (Zhang et al. Citation2019; Shahzadi et al. Citation2020; Yun and Kim Citation2022; Jin et al. Citation2023; Liu et al. Citation2023). The phylogenetic results indicated that S. sagittifolia exhibited the closest relationship with S. polylepis. This study provided new information for the phylogenetic relationship of the Asteraceae family.
Author contributions
L.D. and Z.F. conceived and designed the experiments; X.L., J.L., and T.L. performed the experiments; H.C., T.Q., and B.L. analyzed the data; X.L. and J.L. wrote the manuscript; L.D. and Z.F. revised the manuscript. All authors approved the final version and agreed to be accountable for all aspects of the work.
Ethics statement
This study includes no endangered plant samples, and the sampling site is not located in any protected area. The collection of plant materials is in accordance with local regulations and obtain the permission of local authorities.
Supplemental Material
Download MS Word (309.5 KB)Disclosure statement
No potential conflict of interest was reported by the authors.
Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI at https://www.ncbi.nlm.nih.gov/ under the accession no. ON094066. The associated BioProject, SRA, and Bio-Sample numbers are PRJNA939009, SRR23682417, and SAMN33558826, respectively.
Additional information
Funding
References
- Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, et al. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 19(5):455–477. doi: 10.1089/cmb.2012.0021.
- Doorduin L, Gravendeel B, Lammers Y, Ariyurek Y, Chin-A-Woeng T, Vrieling K. 2011. The complete chloroplast genome of 17 individuals of pest species Jacobaea vulgaris: SNPs, microsatellites and barcoding markers for population and phylogenetic studies. DNA Res. 18(2):93–105. doi: 10.1093/dnares/dsr002.
- Downie SR, Jansen RK. 2015. A comparative analysis of whole plastid genomes from the Apiales: expansion and contraction of the inverted repeat, mitochondrial to plastid transfer of DNA, and identification of highly divergent noncoding regions. Syst Bot. 40(1):336–351. doi: 10.1600/036364415X686620.
- Doyle JJ, Doyle JL. 1987. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull. 19:11–15.
- Hereward JP, Werth JA, Thornby DF, Keenan M, Chauhan BS, Walter GH. 2018. Complete chloroplast genome of glyphosate resistant Sonchus oleraceus L. from Australia, with notes on the small single copy (SSC) region orientation. Mitochondrial DNA B Resour. 3(1):363–364. doi: 10.1080/23802359.2018.1450682.
- Huang L, Wang Z, Wang T, Su YJ. 2016. The complete chloroplast genome sequence of Mikania micrantha (Asteraceae), a noxious invasive weed to South China. Mitochondrial DNA B Resour. 1(1):603–604. doi: 10.1080/23802359.2016.1209090.
- Jin GZ, Li WJ, Song F, Yang L, Wen ZB, Feng Y. 2023. Comparative analysis of complete Artemisia subgenus Seriphidium (Asteraceae: Anthemideae) chloroplast genomes: insights into structural divergence and phylogenetic relationships. BMC Plant Biol. 23(1):136. doi: 10.1186/s12870-023-04113-1.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780. doi: 10.1093/molbev/mst010.
- Li R, Ma PF, Wen J, Yi TS. 2013. Complete sequencing of five Araliaceae chloroplast genomes and the phylogenetic implications. PLOS One. 8(10):e78568. doi: 10.1371/journal.pone.0078568.
- Li T, Xu LS, Chen YS. 2022. Saussurea khunjerabensis (Asteraceae, Cardueae), a new species from Pamir. Phytotaxa. 561(1):65–74. doi: 10.11646/phytotaxa.561.1.6.
- Liu S, Ni Y, Li J, Zhang X, Yang H, Chen H, Liu C. 2023. CPGView: a package for visualizing detailed chloroplast genome structures. Mol Ecol Resour. 23(3):694–704. doi: 10.1111/1755-0998.13729.
- Liu XF, Luo JJ, Zhang MK, Wang Q, Liu J, Wu D, Fu ZX. 2023. Phylogenomic analysis of two species of Parasenecio and comparative analysis within tribe Senecioneae (Asteraceae). Diversity. 15(4):563. doi: 10.3390/d15040563.
- Qu XJ, Moore MJ, Li DZ, Yi TS. 2019. PGA: a software package for rapid, accurate, and flexible batch annotation of plastomes. Plant Methods. 15(1):50. doi: 10.1186/s13007-019-0435-7.
- Seon A, Yun SA, Gil HY, Kim SC. 2017. The complete chloroplast genome sequence of Saussurea polylepis (Asteraceae), a vulnerable endemic species of Korea. Mitochondrial DNA B Resour. 2(2):650–651. doi: 10.1080/23802359.2017.1375881.
- Shahzadi I, Abdullah , Mehmood F, Ali Z, Ahmed I, Mirza B. 2020. Chloroplast genome sequences of Artemisia maritima and Artemisia absinthium: comparative analyses, mutational hotspots in genus Artemisia and phylogeny in family Asteraceae. Genomics. 112(2):1454–1463. doi: 10.1016/j.ygeno.2019.08.016.
- Shi Z, Raab-Straube EV. 2011. Saussurea Candolle. In: Wu ZY, Raven PH, Hong DY, editors. Flora of China. Vols. 20–21. Beijing; St. Louis: Science Press; Missouri Botanical Garden Press; p. 56–149.
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30(9):1312–1313. doi: 10.1093/bioinformatics/btu033.
- Vargas OM, Ortiz EM, Simpson BB. 2017. Conflicting phylogenomic signals reveal a pattern of reticulate evolution in a recent high-Andean diversification (Asteraceae: Astereae: Diplostephium). New Phytol. 214(4):1736–1750. doi: 10.1111/nph.14530.
- Wang AS, Wu HW, Zhu XC, Lin JM. 2018. Species identification of Conyza bonariensis assisted by chloroplast genome sequencing. Front Genet. 9(374):374. doi: 10.3389/fgene.2018.00374.
- Wang B, Zhao Q, Wang XH, Fu ZX. 2020. The complete chloroplast genome of Pertya phylicoides (Asteraceae, Pertyeae): a shurby endemic species from China. Mitochondrial DNA B Resour. 5(1):963–964. doi: 10.1080/23802359.2020.1722763.
- Wang XY, Zhou ZS, Liu G, Qian ZQ. 2018. Characterization of the complete chloroplast genome of the invasive weed Galinsoga quadriradiata (Asterales: Asteraceae). Conserv Genet Resour. 10(1):89–92. doi: 10.1007/s12686-017-0771-8.
- Xu LS, Yi SR, Chen YS. 2020. Saussurea sagittifolia (Asteraceae, Cardueae), a new species from the Bashan Mountains region of China. Phytotaxa. 472(3):295–298. doi: 10.11646/phytotaxa.472.3.9.
- Yun S, Kim S-C. 2022. Comparative plastomes and phylogenetic analysis of seven Korean endemic Saussurea (Asteraceae). BMC Plant Biol. 22(1):550. doi: 10.1186/s12870-022-03946-6.
- Zhang X, Deng T, Moore MJ, Ji YH, Lin N, Zhang HJ, Meng AP, Wang HC, Sun YX, Sun H. 2019. Plastome phylogenomics of Saussurea (Asteraceae: Cardueae). BMC Plant Biol. 19(1):290. doi: 10.1186/s12870-019-1896-6.
- Zhang Y, Zhang J, Yang Y, Liu Q. 2017. Complete chloroplast genome of Pluchea indica (L.) Less. (Asteraceae) and its phylogenetic analysis. Mitochondrial DNA B Resour. 2(2):918–919. doi: 10.1080/23802359.2017.1413299.
