Abstract
Ptomascopus plagiatus (Ménétriés, 1854) is a forensically important silphid species. In this study, we report on the mitochondrial genome of P. plagiatus. The complete mitochondrial genome of P. plagiatus is 17556 bp and contains 22 transfer RNA genes, 13 protein-coding genes (PCGs), two ribosomal RNA genes, and a 2953 bp noncoding region. The nucleotide composition of P. plagiatus is biased toward A and T (A + T: 77.46%). Phylogenetic analysis based on mitogenomic data supports that P. plagiatus is closely related to (Nicrophorus nepalensis Hope, 1831 + Nicrophorus vespilloides Herbst, 1783) within the subfamily Silphinae.
Introduction
Species of subfamily Silphinae are of forensic importance due to their quick colonization of carcasses and relatively long development time (Byrd and Castner Citation2010; Ridgeway et al. Citation2014; Montoya-Molina et al. Citation2021). Ptomascopus plagiatus (Ménétriés, 1854) is a widely distributed silphid species in China (Zheng and Gui Citation1999). Adults and larvae of this species are often collected from buried pork baits at 30 cm depth in Shenyang from late June to late October (Zou et al. Citation2022). Accurate species identification is the prerequisite for the postmortem interval (PMI) estimation using the necrophagous insect succession pattern and development time in forensic entomology (Ren et al. Citation2021). In insect research, the mitochondrial genome plays an important role in phylogenetic inference and species identification (Cameron et al. Citation2007; Shang et al. Citation2019). However, information on the mitochondrial genome of P. plagiatus is lacking, which limits its application in forensic investigations. Therefore, we first provided the complete mitochondrial genome sequence of P. plagiatus in this study.
Materials
Adults of P. plagiatus () were collected using traps baited with pork in Shenyang, Liaoning province, China (41°48′N, 123°25′E). Specimens were identified following the taxonomic keys (Ji Citation2012). The specimens and DNA were stored at the Natural History Museum of Shenyang University (https://museum.syu.edu.cn/, Dianxing Feng, [email protected]) under the voucher number SYU-FD-SIL001.
Figure 1. Adult of Ptomascopus plagiatus (Ménétriés, 1854) (photographed by shutong dai). This species has an orange-red rectangle band on each elytra. The dorsal plate of anterior thorax has dense grayish-yellow appressed hairs. The Middle tibiae are straight or slightly curved, and the hind tibiae are straight. Bar = 1 cm.

Methods
Genomic DNA of muscle tissues from legs and thorax was extracted using a DNeasy Blood & Tissue Kit (Qiagen, Hilden, Germany) according to the manufacturer’s instructions. The DNA was used to construct a library with 350 bp fragments and sequenced using a paired-end strategy 2 × 150 bp on the Novaseq 6000 platform (Illumina, USA) at the commercial corporation (Genepioneer Biotechnologies Co. Ltd., Nanjing, China). The raw reads were filtered using Fastp v0.23.0 (Chen et al. Citation2018). The putative mitochondrial reads were obtained by mapping reads to the local database (including all the published mitochondrial genomes of Coleoptera downloaded from NCBI GenBank database) using Bowtie2 v2.2.6 (Langdon Citation2015). The assembler SPAdes v3.14.1 (Bankevich et al. Citation2012) was used for assembly. To verify the accuracy of the assembly, the coverage depth (3231 × ∼ 7994 ×, mean: 7088.15 ×, Supplementary Figure 1) was computed using Samtools v1.16.1 (Ni et al. Citation2023). Additional, the assembled genome was also aligned with the mitochondrial genome of Nicrophorus nepalensis Hope, 1831 (MW365941) (Cai and Li Citation2021) using Geneious v11.0.4 (Kearse et al. Citation2012). The mitochondrial genome was annotated by MITOS2 online server under the invertebrate mitochondrial code (Bernt et al. Citation2013) and manually verified using the NCBI database. The base composition and codon usage were calculated by MEGA v11.0.13 (Tamura et al. Citation2021). The AT and GC skews were calculated according to the formulae: AT skew = (A − T)/(A + T) and GC skew = (G − C)/(G + C) (Perna and Kocher Citation1995). The final genome map was plotted by CGView online server (Grant and Stothard Citation2008).
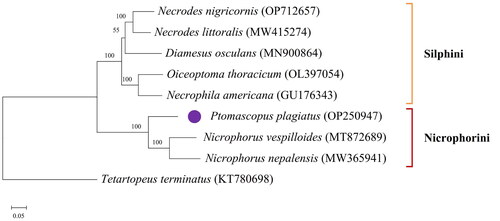
Because the mitochondrial genomes of some silphid species available in the NCBI GenBank database were incomplete and unverified, only the annotated genomes containing at least 13 PCGs and 2 rRNAs sequences were selected for phylogenetic analysis (). The mitochondrial genome of Tetartopeus terminatus (Gravenhorst, 1802) from the subfamily Paederinae was chosen as an outgroup (Kim et al. Citation2023). Sequences were aligned by MAFFT v7.511 (Katoh et al. Citation2002). The GTRGAMMA model was used to construct a ML tree in RAxML v8.2.10 with a bootstrap value of 1000 (Minh et al. Citation2013).
Table 1. List of the mitochondrial genomes used in phylogenetic analysis.
Results
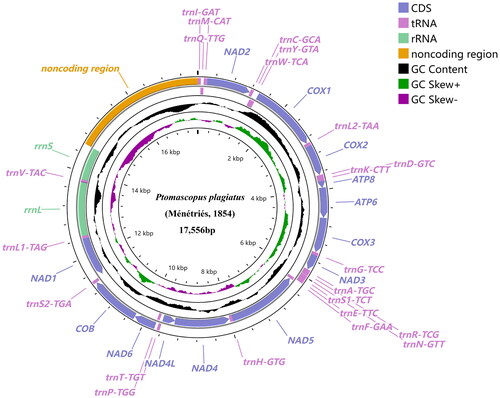
The complete mitochondrial genome of P. plagiatus (GenBank accession no. OP250947) is 17556 bp in length, and contains 22 tRNAs, 13 PCGs, two rRNAs, and one noncoding region which was 2953 bp long (). The nucleotide composition of the mitochondrial genome is 40.38% of A, 37.08% of T, 8.66% of G, and 13.88% of C. The proportion of A + T (77.46%) is higher than G + C (22.54%), and AT skew is positive (0.043). Four PCGs (NAD1, NAD4, NAD4L, and NAD5), eight tRNAs (trnQ, trnC, trnY, trnF, trnH, trnP, trnL1, trnV) and two rRNAs (rrnS, rrnL) are encoded by the light strand, while others are located on the heavy strand (.). Among the 13 PCGs, six genes (COX1, ATP8, NAD2, NAD3, NAD5 and NAD6) begin with ATT codon, five genes (ATP6, NAD4, NAD4L, COX3, and COB) begin with ATG codon, and two genes (COX2 and NAD1) start with ATA codon. Five genes (ATP6, ATP8, NAD2, NAD4L, and NAD6) end with TAA codon, and the rest of the genes except NAD1 (TAG codon) stop with T, the incomplete stop codon. The 13 PCGs encode 3662 codons (excluding stop codons). The four most frequently used codons are UUA, AUU, UUU and AUA, which are used 448, 342, 296, and 230 times, respectively.
Figure 2. Mitochondrial genome map of Ptomascopus plagiatus (Ménétriés, 1854) displaying both heavy (outside the circle) and light (inside the circle) strands. The transcriptional directions of genes located on the heavy and light strands are clockwise and counterclockwise, respectively. Plots of GC content and skew used a sliding window size of 500 and step size of 1. GC skew is plotted using green and purple sliding windows as the deviation from the average of the mitochondrial genome.
The phylogenetic analysis is conducted using 13 PCGs and 2 rRNAs sequences of eight silphid species and one outgroup species. The ML tree shows the eight silphid species cluster together and form the subfamily Silphinae (). There are two groups in this subfamily. In the first group, species from the genera Necrodes, Diamesus, Oiceoptoma and Necrophila are grouped together to form the tribe Silphini. In the other group, P. plagiatus is closely related to the species of genus Nicrophorus, which together constitute the tribe Nicrophorini.
Discussion and conclusion
The circular mitochondrial genome of P. plagiatus is 17556 bp long. Like other typical insect mitochondrial genomes, it contains 13 PCGs, 22 tRNAs, 2 rRNAs, and a noncoding region. The nucleotide composition has obvious A-T bias. Traditionally, Silphidae was a separate family that included two subfamilies, Nicrophorinae and Silphinae. Recently this family was formally downgraded to a subfamily of Staphylinidae (Cai et al. Citation2022; Gruszka and Matuszewski Citation2023; Růžička et al. Citation2023). Correspondingly, Nicrophorinae and Silphinae were relegated to two tribes, Nicrophorini and Silphini, respectively. The molecular phylogenetic analysis supports that P. plagiatus belongs to the tribe Nicrophorini and is close to (N. vespilloides + N. nepalensis) within the subfamily Silphinae. The annotated mitochondrial genome of P. plagiatus provided by this study is extremely valuable for the species identification and phylogenetic analysis of the subfamily Silphinae.
Ethical approval
Ptomascopus plagiatus (Ménétriés, 1854) is a saprophagous species easily trapped using pork baits. The samples collected and used for this study do not involve humans or animals. Therefore, this study does not need ethical approval or permissions to collect the samples.
Authors contributions
WH performed the experiments, analyzed the data and wrote the original manuscript. DF was involved in the conception, design and revising the manuscript. SD collected and photographed the specimen, and analyzed the data. All authors have approved the manuscript for publication and agreed to be accountable for all aspects of the work.
Supplemental Material
Download Text (117.2 KB)Supplemental Material
Download MS Word (93 KB)Disclosure statement
No potential conflict of interest is reported by the authors.
Data availability statement
Mitogenome data supporting this study are openly available in GenBank at nucleotide database, https://www.ncbi.nlm.nih.gov/nuccore/OP250947.1. The associated BioProject, SRA, and BioSample numbers are PRJNA876787, SRR21634603, and SAMN30815897, respectively.
Additional information
Funding
References
- Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, et al. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 19(5):455–477. doi: 10.1089/cmb.2012.0021.
- Bernt M, Donath A, Jühling F, Externbrink F, Florentz C, Fritzsch G, Pütz J, Middendorf M, Stadler PF. 2013. MITOS: improved de novo metazoan mitochondrial genome annotation. Mol Phylogenet Evol. 69(2):313–319. doi: 10.1016/j.ympev.2012.08.023.
- Byrd JH, Castner JL. 2010. Forensic entomology: the utility of arthropods in legal investigations. 2nd ed. Boca Raton: CRC Press.
- Cai Y, Li X. 2021. The complete mitochondrial genome of a burying beetle, Nicrophorus nepalensis Hope, 1831 (Coleoptera: silphidae). Mitochondrial DNA B Resour. 6(6):1727–1728. doi: 10.1080/23802359.2021.1930220.
- Cai C, Tihelka E, Giacomelli M, Lawrence JF, Ślipiński A, Kundrata R, Yamamoto S, Thayer MK, Newton AF, Leschen RAB, et al. 2022. Integrated phylogenomics and fossil data illuminate the evolution of beetles. R Soc Open Sci. 9(3):211771. doi: 10.1098/rsos.211771.
- Cameron SL, Lambkin CL, Barker SC, Whiting MF. 2007. A mitochondrial genome phylogeny of Diptera: whole genome sequence data accurately resolve relationships over broad timescales with high precision. Syst Entomol. 32(1):40–59. doi: 10.1111/j.1365-3113.2006.00355.x.
- Chen S, Zhou Y, Chen Y, Gu J. 2018. fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics. 34(17):i884–i890. doi: 10.1093/bioinformatics/bty560.
- Grant JR, Stothard P. 2008. The CGView Server: a comparative genomics tool for circular genomes. Nucleic Acids Res. 36(Web Server issue):W181–184. doi: 10.1093/nar/gkn179.
- Gruszka J, Matuszewski S. 2023. Initial laboratory validation of temperature development models for Necrodes littoralis L. (Staphylinidae: silphinae). Int J Legal Med. 137(3):903–911. doi: 10.1007/s00414-023-02969-4.
- Ji Y. 2012. The carrion beetles of China (Coleopyera: silphidae). Beijing, China: China Forestry Press.
- Jiang Y, Liu Z, Pi Z, Xie Q, Meng F, Cai J. 2021. The complete mitochondrial genome of a corpse related necrophagous beetle, Necrodes littoralis (Coleoptera: silphidae). Mitochondrial DNA B Resour. 6(4):1512–1513. doi: 10.1080/23802359.2021.1914236.
- Katoh K, Misawa K, Kuma K, Miyata T. 2002. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 30(14):3059–3066. doi: 10.1093/nar/gkf436.
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, et al. 2012. Geneious basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 28(12):1647–1649. doi: 10.1093/bioinformatics/bts199.
- Kim MI, Park JS, Choung CM, Kim MJ, Kim I. 2023. Complete mitochondrial genome of the forensically important carrion beetle, Necrodes nigricornis (Coleoptera: silphidae). J Asia-Pac Entomol. 26(1):102033. doi: 10.1016/j.aspen.2022.102033.
- Langdon WB. 2015. Performance of genetic programming optimised Bowtie2 on genome comparison and analytic testing (GCAT) benchmarks. BioData Min. 8(1)
- Minh BQ, Nguyen MAT, von Haeseler A. 2013. Ultrafast approximation for phylogenetic bootstrap. Mol Biol Evol. 30(5):1188–1195. doi: 10.1093/molbev/mst024.
- Montoya-Molina S, Jakubec P, Qubaiová J, Novák M, Šuláková H, Růžička J. 2021. Developmental models of the carrion beetle Thanatophilus rugosus (Linnaeus, 1758) (Coleoptera: silphidae). Sci Rep. 11(1):19377. doi: 10.1038/s41598-021-98833-9.
- Ni Y, Li JL, Zhang C, Liu C. 2023. Generating sequencing depth and coverage map for organelle genomes. protocols.io. doi: 10.17504/protocols.io.4r3l27jkxg1y/v1.
- Perna NT, Kocher TD. 1995. Patterns of nucleotide composition at fourfold degenerate sites of animal mitochondrial genomes. J Mol Evol. 41(3):353–358. doi: 10.1007/BF01215182.
- Ren LP, Shang YJ, Guo YD. 2021. Progress and application of entomological evidence in forensic science. Fa Yi Xue Za Zhi. 37:295.
- Ridgeway JA, Midgley JM, Collett IJ, Villet MH. 2014. Advantages of using development models of the carrion beetles Thanatophilus micans (Fabricius) and T. mutilatus (Castelneau) (Coleoptera: silphidae) for estimating minimum post mortem intervals, verified with case data. Int J Legal Med. 128(1):207–220. doi: 10.1007/s00414-013-0865-0.
- Růžička J, Jakubec P, Mahlerová K, Šípková H, Nishikawa M. 2023. Integrative taxonomy and species distribution models of the genus Diamesus Hope, 1840 (Coleoptera: staphylinidae: silphinae). Sci Rep. 13(1):3192. doi: 10.1038/s41598-023-30019-x.
- Shang Y, Ren L, Chen W, Zha L, Cai J, Dong J, Guo Y. 2019. Comparative mitogenomic analysis of forensically important sarcophagid flies (Diptera: sarcophagidae) and implications of species identification. J Med Entomol. 56(2):392–407. doi: 10.1093/jme/tjy162.
- Song H, Sheffield NC, Cameron SL, Miller CK, Whiting MF. 2010. When phylogenetic assumptions are violated: base compositional heterogeneity and among-site rate variation in beetle mitochondrial phylogenomics. Syst Entomol. 35(3):429–448. doi: 10.1111/j.1365-3113.2009.00517.x.
- Tamura K, Stecher G, Kumar S. 2021. MEGA 11: molecular evolutionary genetics analysis version 11. Mol Biol Evol. 38(7):3022–3027. doi: 10.1093/molbev/msab120.
- Zhang X, Hou Q, Zhang L, Cai J, Meng F. 2020. The complete mitochondrial genome of a potentially forensic related carrion beetle, Diamesus osculans (Vigors, 1825). Mitochondrial DNA B Resour. 5(2):1423–1424. doi: 10.1080/23802359.2020.1736955.
- Zheng LY, Gui H. 1999. Insect taxonomy. Nanjing, China: Nanjing Normal University Press.
- Zou TL, Feng DX, Huang GY, Sun DP, Dai ST. 2022. Species composition and succession of necrophagous insects on small buried baits in China. J Med Entomol. 59(4):1182–1190. doi: 10.1093/jme/tjac045.