Abstract
A recently published complete mitochondrial genome of Spotted Greenshank (Tringa guttifer) was the first DNA sequence of this species (GenBank accession number MK905885, RefSeq number NC_044665; Liu et al. Citation2019, The complete mitochondrial genome of the Spotted Greenshank Tringa guttifer (Charadriiforemes [sic]: Charadriidae), Mitochondrial DNA Part B. 4:2353–2354). Here we show that this mitogenome is actually a chimera containing DNA fragments of both a Tringa sandpiper (presumably T. guttifer) and the Red-necked Stint (Calidris ruficollis). This mitogenome has been re-used in at least three phylogenies. The error is documented to avoid the perpetuation of erroneous sequence information in the literature.
Introduction
Spotted Greenshank Tringa guttifer (Nordmann, 1835) is an endangered shorebird (Charadriiformes) breeding in northeastern Asia and wintering in southeast Asia. The first published DNA sequence of this species was a complete mitochondrial genome (hereafter mitogenome) published by Liu et al. (Citation2019). This sequence was derived from a sample taken from an individual captured along the eastern coast of Jiangsu province, China (GenBank accession number MK905885, RefSeq number NC_044665). Liu et al. (Citation2019) included a phylogram based on complete mitogenomes which placed the T. guttifer sequence among other members of the genus Tringa. Using a strategy described by Norén and Kullander (Citation2018), we show that accession MK905885 is actually a chimera containing DNA of two species of shorebirds.
Materials and methods
We verified the identity of MK905885 by performing separate phylogenetic analyses of each of the two ribosomal RNA markers and the 12 protein-coding genes and comparing the position of each species in the gene trees: 12S ribosomal RNA (12S rRNA, 972 bp), 16S ribosomal RNA (16S rRNA, 1582 bp), NADH dehydrogenase subunit 1 (ND1, 971 bp), NADH dehydrogenase subunit 2 (ND2, 1041 bp), cytochrome oxidase subunit I (COI, 1551 bp), cytochrome oxidase subunit II (COII, 684 bp), Adenosine Tri-Phosphate 8 and 6 (ATP8-6, 844 bp), cytochrome oxidase subunit III (COIII, 784 bp), NADH dehydrogenase subunit 3 (ND3, 349 bp), NADH dehydrogenase subunit 4 (ND4, 1668 bp), NADH dehydrogenase subunit 5 (ND5, 1815 bp), cytochrome b (cyt b, 1143 bp) and NADH dehydrogenase subunit 6 (ND6, 525 bp). We included all species of Tringa and Calidris for which mitogenomes were available at the time of writing, plus relevant outgroups, based on a previous study of the relationships of shorebirds (Gibson and Baker Citation2012). We excluded a problematic sequence of T. totanus (MK922124/NC_044648) (see Sangster and Luksenburg Citation2021a). The MITOS2 web server (Bernt et al. Citation2013) was used to obtain information on the first and last positions of individual genes. CLUSTALW (as implemented in MEGA7, Kumar et al. Citation2016) was used to align sequences.
When we noticed that one of these data sets showed a different, but strongly supported, position of T. guttifer than the other data sets we visually compared the T. guttifer sequence with those of other shorebirds and determined the first and last positions of the anomalous fragment (Sangster and Luksenburg Citation2021a). We constructed separate phylogenies of (i) the anomalous fragment (953 bp) and (ii) the rest of the mitogenome. The latter phylogeny was constructed with the data set trimmed by GBLOCKS (Castresana Citation2000). GBLOCKS eliminates poorly aligned positions and divergent regions, which may not be homologous or may have been saturated by multiple substitutions (Castresana Citation2000). This resulted in an alignment of 14,346 bp. Maximum Likelihood phylogenies were obtained using MEGA7. The appropriate substitution model for each data set was selected using the Akaike Information Criterion. The selected models were GTR + G and GTR + G + I, respectively. Sequence divergence was calculated as uncorrected p-values with complete deletion of nucleotide positions with missing data.
Results
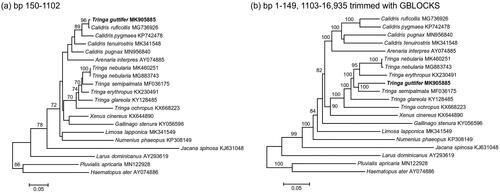
Initial analysis, based on gene trees of each mitochondrial gene, showed that in the 12s rRNA gene tree, T. guttifer clustered with Red-necked Stint Calidris ruficollis, with strong support, whereas the other gene trees placed T. guttifer among members of the genus Tringa. Direct (visual) comparison of the mitogenome sequences showed that the anomalous part consisted of a single 953 bp fragment, located at positions 150–1102. This represented 5.7% of the total length of the published mitogenome of T. guttifer (16,835 bp, not 16,935 bp as stated in Liu et al. Citation2019). A Maximum Likelihood (ML) phylogeny of this portion of the mitogenome is shown in , which shows a sister-relationship of the mitogenomes of T. guttifer and C. ruficollis with 96% bootstrap support. Sequence divergence between this portion of the mitogenomes of T. guttifer and C. ruficollis was minimal (0.5%). In contrast, sequence divergence between this portion of the mitogenomes of T. guttifer and the other species of Tringa included in this study ranged from 7.2% to 9.0%. Sequence divergence between this portion of the mitogenomes of species of Tringa ranged from 3.1% to 8.7%. A ML phylogeny of the other part of the mitogenome is shown in , which placed T. guttifer as the sister of T. semipalmata with 100% bootstrap support.
Figure 1. ML phylogenies of shorebirds (Charadriiformes) based on (a) positions 150–1102 (953 bp) of the mitogenome, (b) mitogenomes excluding positions 150–1102 and trimmed with GBLOCKS (14,346 bp). Numbers along branches represent bootstrap support values (>70%) based on 1000 pseudoreplications. Note the different position of T. guttifer in the two gene trees.
Discussion
Our results show that different parts of the published mitogenome of T. guttifer cluster with different species, each with strong bootstrap support. One of the fragments was sister to that of C. ruficollis, a smaller species of shorebird. Phylogenetic studies have shown that C. ruficollis is the sister species of C. pygmaea, and that all other species of Calidris are more distantly related from these two species (Gibson and Baker Citation2012; Černý and Natale Citation2022). Our study did not include all species of Calidris but because we included both C. ruficollis and C. pygmaea, and the 953 bp fragment, located at positions 150–1102, of the published mitogenome of T. guttifer clustered with C. ruficollis with strong support, we are confident that this fragment is correctly identified as belonging to the latter species. Because no previously published mitochondrial sequences were available of T. guttifer, assessing the identity of the other part of the mitogenome was not possible but it may well have been of T. guttifer.
The mitogenome of T. guttifer was obtained with Sanger sequencing. The chimera likely occured in the laboratory resulting from the transfer of a sample of C. ruficollis to a tube intended for T. guttifer before PCR amplification or before DNA sequencing. Indeed, a mitogenome of C. ruficollis was sequenced by members of the same team and was published in the same year as that of T. guttifer (Chen et al. Citation2019). Detecting such errors is possible if each fragment is separately analyzed phylogenetically before assembling the fragments into a single mitogenome.
Sangster and Luksenburg (Citation2021a) used three markers commonly-used in ornithology (ND2, COI, cyt b) to verify the identity of 1559 mitogenomes of birds. They found 78 problematic mitogenomes, including 23 chimeras, but noted that this must represent an underestimate of the true prevalence of problematic mitogenomes because the three markers only represent a small portion of the mitogenome. The present study adds another chimera to this set and shows that using other markers indeed reveals additional problematic sequences.
As noted previously, reporting problematic mitogenomes is necessary because accumulation of erroneous sequences may compromise subsequent applications, including DNA identification, primer design for intraspecific studies, phylogenetic inference, historical biogeography, taxonomy and comparative analysis (Sangster and Luksenburg Citation2021b). Indeed, we found three re-uses of the mitogenome (Guo et al. Citation2021; Yang et al. Citation2021; Černý and Natale Citation2022). In each case, the mitogenome (or parts thereof) was included in a phylogeny.
Our study underscores that chimeras are easily overlooked without dedicated analysis. We suspect that the few cases of chimerism reported so far in vertebrate mitogenomics (e.g. Norén and Kullander Citation2018; Sangster and Luksenburg Citation2020, Citation2021a, Citation2021c) do not reflect the true prevalence of this problem. Clearly, greater vigilance is necessary during laboratory procedures, quality control of raw data and peer review of the final sequences.
Authors’ contributions
GS conceived the study, performed the analyses and wrote the first draft. JAL helped interpret the results and revised the manuscript for intellectual content. Both authors approved the version to be published and agree to be accountable for all aspects of the work.
Acknowledgments
We are grateful to the two referees for providing constructing comments that helped us improve the clarity of our paper.
Disclosure statement
No potential conflict of interest was reported by the authors.
Data availability statement
The paper we comment on is available at https://doi.org/10.1080/23802359.2019.1629349. The sequence data that support the findings of this study were published previously and are openly available on GenBank at https://www.ncbi.nlm.nih.gov/nucleotide. The mitogenome of Tringa guttifer is available at https://www.ncbi.nlm.nih.gov/nuccore/MK905885.
Additional information
Funding
References
- Bernt M, Donath A, Jühling F, Externbrink F, Florentz C, Fritzsch G, Pütz J, Middendorf M, Stadler PF. 2013. MITOS: improved de novo metazoan mitochondrial genome annotation. Mol Phylogenet Evol. 69(2):313–319. doi:10.1016/j.ympev.2012.08.023.
- Castresana J. 2000. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol Biol Evol. 17(4):540–552. doi:10.1093/oxfordjournals.molbev.a026334.
- Černý D, Natale R. 2022. Comprehensive taxon sampling and vetted fossils help clarify the time tree of shorebirds (Aves, Charadriiformes). Mol Phylogenet Evol. 177:107620. doi:10.1016/j.ympev.2022.107620.
- Chen W, Liu W, Zhang C, Li K, Hu C, Chang Q. 2019. The complete mitochondrial genome of the Red-necked Stint Calidris ruficollis (Charadriiformes, Scolopacidae). Conservation Genet Resour. 11(2):181–184. doi:10.1007/s12686-018-0996-1.
- Gibson R, Baker A. 2012. Multiple gene sequences resolve phylogenetic relationships in the shorebird suborder Scolopaci (Aves: Charadriiformes). Mol Phylogenet Evol. 64(1):66–72. doi:10.1016/j.ympev.2012.03.008.
- Guo T, Yang Z, Zhu Y, Wang H, Kong D, Hu C, Liu M. 2021. The complete mitochondrial genome of Eastern Curlew Numenius madagascariensis (Charadriiformes, Scolopacidae). Mitochondrial DNA B Resour. 6(11):3091–3092. doi:10.1080/23802359.2021.1959448.
- Kumar S, Stecher G, Tamura K. 2016. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 33(7):1870–1874. doi:10.1093/molbev/msw054.
- Liu W, He Y, Ding J, Chang Q. 2019. The complete mitochondrial genome of the Spotted Greenshank Tringa guttifer (Charadriiforemes [sic]: Charadriidae). Mitochondrial DNA B Resour. 4(2):2353–2354. doi:10.1080/23802359.2019.1629349.
- Norén M, Kullander S. 2018. The enigmatic Betadevario ramachandrani (Teleostei: Cyprinidae: Danioninae): phylogenetic position resolved by mitogenome analysis, with remarks on the prevalence of chimeric mitogenomes in GenBank. Cogent Biol. 4(1):1525857. doi:10.1080/23312025.2018.1525857.
- Sangster G, Luksenburg JA. 2020. The published complete mitochondrial genome of Eptesicus serotinus is a chimera of Vespertilio sinensis and Hypsugo alaschanicus (Mammalia: Chiroptera). Mitochondrial DNA B Resour. 5(3):2661–2664. doi:10.1080/23802359.2020.1785349.
- Sangster G, Luksenburg JA. 2021a. Sharp increase of problematic mitogenomes of birds: causes, effects and remedies. Gen Biol Evol. 13:evab210. doi:10.1093/gbe/evab210.
- Sangster G, Luksenburg JA. 2021b. The published complete mitochondrial genome of milk shark (Rhizoprionodon acutus) is a misidentified Pacific spadenose shark (Scoliodon macrorhynchos) (Chondrichthyes: Carcharhiniformes). Mitochondrial DNA B Resour. 6(3):828–830. doi:10.1080/23802359.2021.1884019.
- Sangster G, Luksenburg JA. 2021c. Scientific data laundering: chimeric mitogenomes of a sparrowhawk and a nightjar covered-up by forged phylogenies. Biochem Syst Ecol. 96:104263. doi:10.1016/j.bse.2021.104263.
- Yang C, Hou X, Liu B-Y, Gong H-S, Yuan H, Li X-J, Tang J, Wang Y. 2021. The mitogenome of Common Snipe, Gallinago gallinago gallinago Linnaeus, 1758 and evolutionary implications for the family Scolopacidae. Mitochondrial DNA B Resour. 6(10):2886–2889. doi:10.1080/23802359.2021.1972870.
