Abstract
Ophiuroids are a diversified benthic taxon in the deep sea. Given their various dispersal strategies, they are considered an adequate group to assess genetic connectivity, especially in the seamounts that function as islands. Ophioleila elegans A.H. Clark, 1949, in the family Ophiothamnidae, was previously reported from the Caiwei Guyot, a seamount in the northwest Pacific Ocean. Here, we described the mitochondrial genome of O. elegans collected from another seamount in the northwest Pacific. The whole mitogenome is 16,376 bp in length and encodes 13 protein-coding genes, two ribosomal RNA genes, and 22 transfer RNA genes. Phylogenetic analysis based on the mitogenome sequences showed that O. elegans was clustered with Histampica sp., the only species for which mitogenome sequence has been reported within the family Ophiothamnidae. The complete mitogenome of O. elegans first reported in the present study provides useful information for population genetics and evolutionary relationship of this taxon, especially in the northwest Pacific seamounts.
Introduction
Ophiuroids are a particularly represented taxon in most deep-sea benthic habitats and have various strategies for dispersal (O'Hara Citation2007). Given their abundance and diverse dispersal strategies, they are considered an adequate group to assess genetic connectivity dynamics in seamounts that serve as centers of endemism and stepping stones for dispersal (Cho and Shank Citation2010; Na et al. Citation2021). Ophioleila elegans A.H. Clark, 1949 is a rarely reported ophiuroid species distributed from the north-central Pacific Ocean to the northwest Pacific seamounts (Zhang et al. Citation2018). In this respect, O. elegans seems to be the appropriate species for evaluating genetic connectivity of seamounts, especially in the northwest Pacific Ocean. Here, we described the complete mitochondrial genome of O. elegans and analyzed its phylogenetic relationship with mitogenomes of other ophiuroid species.
Materials
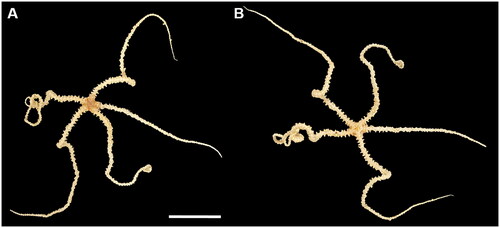
During the oceanographic cruise WP21 of the Korea Institute of Ocean Science and Technology (KIOST) in May 2021, the ophiuroid specimens were collected from the Shkolnik Guyot, northwest Pacific Ocean (16° 5′ 45″ N, 151° 53′ 6.34″ E; depth: 1474 m) using ROV URI-L (Redone Technologies, Jangseong, Republic of Korea). Photographs of the specimens were taken on-board with a Nikon DSLR (). The specimens were identified as Ophioleila elegans by the following morphological characteristics: oral shields small and triangular, adoral shields enlarged trapezoid shape, five arms more than five times the disc diameter (Zhang et al. Citation2018). The O. elegans specimens were deposited at the Marine Biodiversity Institute of Korea (https://www.mabik.re.kr/html/en/, Seok-Chun Ko, and [email protected]) under the voucher number MABIK BK20220511.
Methods
Genomic DNA was extracted from the specimen using DNeasy Blood & Tissue Kit (Qiagen, Hilden, Germany). The mitochondrial cox1 region of the specimen was confirmed, which showed 100% sequence identity to O. elegans (accession number: KU895364). For the mitochondrial genome analysis, mitochondrial DNA amplification was performed using REPLI-g Mitochondrial DNA kit (Qiagen, Hilden, Germany). The REPLI-g kit was used to generate high yields of enriched mitochondrial DNA, which has already been applied to ophiuroids (Lee et al. Citation2019). Sequencing libraries were prepared using the TruSeq Nano DNA High Throughput Library Prep Kit (Illumina, San Diego, CA). Genomic DNA of 100 ng was sheared using adaptive focused acoustic technology (Covaris, Woburn, MA) and the fragmented DNA was end-repaired to create 5′-phosphorylated, blunt-ended dsDNA molecules. After end-repair, the DNA was size-selected using a bead-based method. These DNA fragments go through single ‘A’ base addition and ligation to the TruSeq DNA UD Indexing adapters. The products are then purified and enriched with PCR to generate the final DNA library. The paired end (2 × 150 bp) sequencing was performed through Illumina NovaSeq platform (Illumina, San Diego, CA). The low-quality reads and adapter sequences were trimmed by Trimmomatic v0.36 (Bolger et al. Citation2014), and the trimmed reads were further assembled using SPAdes 3.15.0 (Bankevich et al. Citation2012). The overlapping region was cut out, the remaining sequence was considered circular, and oriented rotation was performed. The closing of the circle was verified by Sanger sequencing. The assembled genome was annotated using MITOS2 (Donath et al. Citation2019) and these annotations were manually adjusted by comparison with other ophiuroid mitogenomes. The mitochondrial genome map and the sequencing depth and coverage map were described using the Proksee (Grant et al. Citation2023) and CircularMapper (Peltzer et al. Citation2016; Fig. S1), respectively. Since the genus Ophioleila is monotypic, and only one mitogenome sequence (Histampica sp. Am Clark, 1970; MZ442339) has been reported within the family Ophiothamnidae, eight mitogenome sequences belonging to the suborder Gnathophiurina were added for phylogenetic analysis, together with three outgroup species (). We aligned the 13 protein-coding genes (PCGs) using MAFFT v7.49 (Katoh and Standley Citation2013) and selected the evolutionary best-substitution model for each gene through IQ-TREE webserver (Trifinopoulos et al. Citation2016). A phylogenetic tree was reconstructed by the maximum-likelihood (ML) method with the 1000 bootstrap replicates and the Bayesian inference (BI) method using RAxML-NG v1.2.0 and MrBayes v3.2.7, respectively (Ronquist et al. Citation2012; Kozlov et al. Citation2019). The bootstrap support (BS) values from the ML and the Bayesian posterior probability (BPP) from the BI were indicated in each node. The phylogenetic tree based on the ML algorithm was visualized using FigTree v1.4.4 (http://tree.bio.ed.ac.uk/software/figtree).
Table 1. Mitochondrial genomes belonging to the suborder Gnathophiurina and three outgroup species used for phylogenetic analysis.
Results
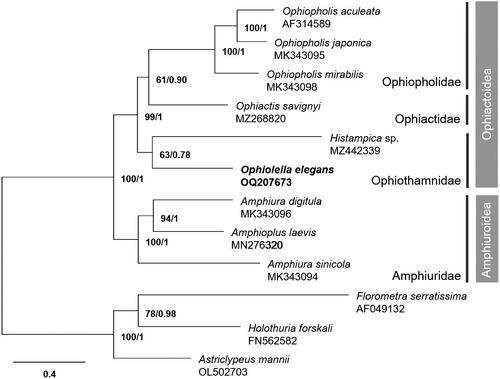
The whole mitogenome of O. elegans is 16,376 bp in length (GenBank accession no. OQ207673), with 64% and 36% of AT and GC contents, respectively (). It consists of 13 PCGs, two ribosomal RNA genes (rRNAs), and 22 transfer RNA genes (tRNAs). All PCGs have ATG as the start codon and TAA as the stop codon, except cytb and cox1, which start with GTG. Phylogenetic analysis based on 13 PCGs shows that O. elegans is most closely related to H. sp. (MZ442339), but with weak support (BS: 63, BPP: 0.78; ).
Figure 2. Mitochondrial genome map of Ophioleila elegans. GC content was plotted by default values of window size (500) and score scale (1).
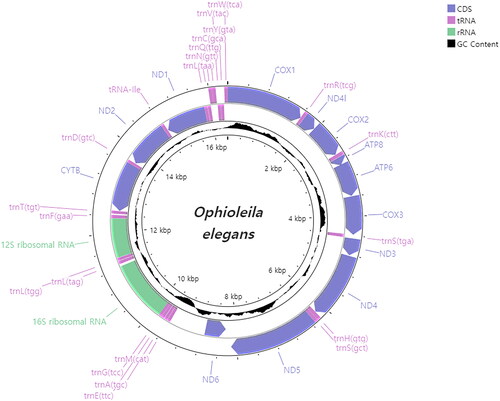
Figure 3. The maximum-likelihood (ML) phylogenetic tree of Ophioleila elegans and other ophiuroid species based on 13 protein-coding mitochondrial genes. Numbers on each node denote the bootstrap support values from the ML and the Bayesian posterior probabilities from the Bayesian inference (BI) method. The scale bar indicates the number of substitutions per site.
Discussion and conclusions
The conserved tRNA cluster (Asn-Gln-Cys-Val-Tyr-Trp) commonly identified in the order Amphilepidida, to which O. elegans belongs (Lee et al. Citation2019), was also found in this study. In the phylogenetic analysis, O. elegans and H. sp. were clustered with low BS and BPP values. Ophioleila was tentatively placed in the family Ophiothamnidae with the genera Ophiothamnus and Histampica based on the exon-capture phylogeny (O'Hara et al. Citation2017; Zhang et al. Citation2018). However, the low support value in the phylogenetic analysis between O. elegans and Ophiothamnus species has also been reported in previous study (Zhang et al. Citation2018). Therefore, the additional mitogenome data should be supplemented to better understand the phylogenetic relationships of the family Ophiothamnidae. The complete mitogenome of O. elegans first reported in the present study would provide helpful information for future studies on evolutionary relationships of this deep-sea taxon.
Author contributions
CP: writing the original manuscript, experiment, and data analysis; EK: data analysis and revising the manuscript; SJ: conceptualization and revising the manuscript. All authors approved the final version of the manuscript.
Ethical approval
No ethical approval was required for this study.
Supplemental Material
Download MS Word (298.2 KB)Supplemental Material
Download PNG Image (282 KB)Disclosure statement
The authors report that there are no conflicts of interest.
Data availability statement
The mitogenome sequence data that support the findings of this study are openly available in GenBank of NCBI at https://www.ncbi.nlm.nih.gov/ under the accession no. OQ207673. The associated BioProject, SRA, and Bio-Sample numbers are PRJNA943565, SRR23824997, and SAMN33731640, respectively.
Additional information
Funding
References
- Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, et al. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 19(5):455–477. doi: 10.1089/cmb.2012.0021.
- Bolger AM, Lohse M, Usadel B. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 30(15):2114–2120. doi: 10.1093/bioinformatics/btu170.
- Cho W, Shank TM. 2010. Incongruent patterns of genetic connectivity among four ophiuroid species with differing coral host specificity on North Atlantic seamounts. Mar Ecol. 31(Suppl. 1):121–143. doi: 10.1111/j.1439-0485.2010.00395.x.
- Donath A, Juhling F, Al-Arab M, Bernhart SH, Reinhardt F, Stadler PF, Middendorf M, Bernt M. 2019. Improved annotation of protein-coding genes boundaries in metazoan mitochondrial genomes. Nucleic Acids Res. 47(20):10543–10552. doi: 10.1093/nar/gkz833.
- Grant JR, Enns E, Marinier E, Mandal A, Herman EK, Chen CY, Graham M, Van Domselaar G, Stothard P. 2023. Proksee: in-depth characterization and visualization of bacterial genomes. Nucleic Acids Res. 51(W1):W484–W492. doi: 10.1093/nar/gkad326.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780. doi: 10.1093/molbev/mst010.
- Kozlov AM, Darriba D, Flouri T, Morel B, Stamatakis A. 2019. RAxML-NG: a fast, scalable and user-friendly tool for maximum likelihood phylogenetic inference. Bioinformatics. 35(21):4453–4455. doi: 10.1093/bioinformatics/btz305.
- Lee T, Bae YJ, Shin S. 2019. Mitochondrial gene rearrangement and phylogenetic relationships in the Amphilepidida and Ophiacanthida (Echinodermata, Ophiuroidea). Mar Biol Res. 15(1):26–35. doi: 10.1080/17451000.2019.1601226.
- Li QH, Li YX, Na JY, Han XQ, Paterson GLJ, Liu K, Zhang DS, Qiu JW. 2021. Description of a new species of Histampica (Ophiuroidea: Ophiothamnidae) from cold seeps in the South China Sea and analysis of its mitochondrial genome. Deep Sea Res I Oceanogr Res Pap. 178:103658. doi: 10.1016/j.dsr.2021.103658.
- Na JY, Chen WY, Zhang DS, Zhang RY, Lu B, Shen CC, Zhou YD, Wang CS. 2021. Morphological description and population structure of an ophiuroid species from cobalt-rich crust seamounts in the Northwest Pacific: implications for marine protection under deep-sea mining. Acta Oceanol Sin. 40(12):79–89. doi: 10.1007/s13131-020-1666-1.
- O'Hara TD, Hugall AF, Thuy B, Stöhr S, Martynov AV. 2017. Restructuring higher taxonomy using broad-scale phylogenomics: the living Ophiuroidea. Mol Phylogenet Evol. 107:415–430. doi: 10.1016/j.ympev.2016.12.006.
- O'Hara TD. 2007. Seamounts: centres of endemism or species richness for ophiuroids? Global Ecol Biogeogr. 16(6):720–732.
- Peltzer A, Jäger G, Herbig A, Seitz A, Kniep C, Krause J, Nieselt K. 2016. EAGER: efficient ancient genome reconstruction. Genome Biol. 17(1):60. doi: 10.1186/s13059-016-0918-z.
- Perseke M, Bernhard D, Fritzsch G, Brummer F, Stadler PF, Schlegel M. 2010. Mitochondrial genome evolution in Ophiuroidea, Echinoidea, and Holothuroidea: insights in phylogenetic relationships of Echinodermata. Mol Phylogenet Evol. 56(1):201–211. doi: 10.1016/j.ympev.2010.01.035.
- Ronquist F, Teslenko M, van der Mark P, Ayres DL, Darling A, Hohna S, Larget B, Liu L, Suchard MA, Huelsenbeck JP. 2012. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol. 61(3):539–542. doi: 10.1093/sysbio/sys029.
- Scouras A, Smith MJ. 2001. A novel mitochondrial gene order in the crinoid echinoderm Florometra serratissima. Mol Biol Evol. 18(1):61–73. doi: 10.1093/oxfordjournals.molbev.a003720.
- Shin JS, Song CU, Choi H, Kwon KK, Kang D, Eyun SI, Choi KS. 2022. The complete mitochondrial genome of the sand dollar Astriclypeus mannii (Verrill, 1867) (Echinoidea: Astriclypeidae) in the subtidal sand flat in Jeju Island off the south coast of Korea. Mitochondrial DNA B Resour. 7(9):1602–1603. doi: 10.1080/23802359.2022.2116946.
- Smith MJ, Arndt A, Gorski S, Fajber E. 1993. The phylogeny of echinoderm classes based on mitochondrial gene arrangements. J Mol Evol. 36(6):545–554. doi: 10.1007/BF00556359.
- Trifinopoulos J, Nguyen LT, von Haeseler A, Minh BQ. 2016. W-IQ-TREE: a fast online phylogenetic tool for maximum likelihood analysis. Nucleic Acids Res. 44(W1):W232–W235. doi: 10.1093/nar/gkw256.
- Xu QZ, Li YX, Dong Y. 2019. Characterization of the complete mitochondrial genome of Amphioplus laevis (Ophiuroidea, Amphiuridae) with phylogenetic analysis. Mitochondrial DNA B Resour. 4(2):3062–3063. doi: 10.1080/23802359.2019.1667907.
- Zhang D, Lu B, Wang C, O’Hara TD. 2018. The first record of Ophioleila elegans (Echinodermata: Ophiuroidea) from a deep-sea seamount in the Northwest Pacific Ocean. Acta Oceanol Sin. 37(10):180–184. doi: 10.1007/s13131-018-1323-0.