Abstract
Tetraselmis marina (Cienkowski) R.E.Norris, Hori & Chihara 1980, a costal green microalga, is considered as a promising animal feed in aquaculture due to the high content of fatty acids and carotenoid. Furthermore, T. marina plays important roles in bioremediation. In this study, we assembled the complete chloroplast genome of T. marina. Results showed that the full length of the complete chloroplast genome was 96,151 bp, containing a large single-copy region of 62,574 bp, a small single-copy region of 1261 bp, and a pair of inverted repeat regions of 16,158 bp. The GC content of the genome was 36.6%. A total of 125 genes were annotated, including 81 protein coding genes, 38 tRNA genes, and six rRNA genes. Phylogenetic analysis based on 22 chloroplast genomes suggested that T. marina was closely related to Tetraselmis sp. CCMP 881.
1. Introduction
Tetraselmis marina (Cienkowski) R.E.Norris, Hori & Chihara 1980, a common coastal microalga, belongs to genus Tetraselmis (Chlorodendraceae). Tetraselmis sp. is commonly considered as a promising potential source of antioxidants or animal feed due to the high contents of fatty acid and carotenoid profile (Moussa et al. Citation2017; Oliveira Moser et al. Citation2022). Additionally, several species of Tetraselmis sp. are commonly potential candidates for radioactive Sr bioremediation (Fukuda et al. Citation2014). Interestingly, several species of Tetraselmis sp. possess biomineralization capacity, and can produce intracellular inclusions of amorphous calcium carbonate (i.e. micropearls, Martignier et al. Citation2018, Citation2020). It implies that Tetraselmis sp. may play a role in the ocean carbon cycle. However, the information about the chloroplast genome of T. marina has been not reported. In the present study, we sequenced and assembled the chloroplast genome of T. marina, and analyzed its phylogenetic position.
2. Materials
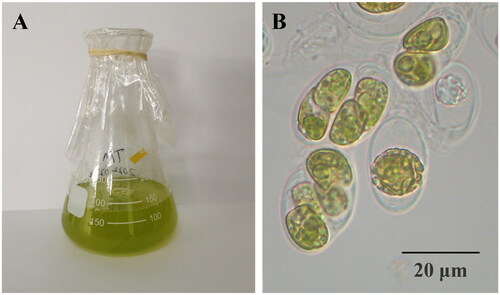
T. marina was isolated from coral Pocillopora damicornis in the coast of Sanya City, China (18°18′ N and 109°48′ E), and cultured in f/2 medium (). It was deposited at the laboratory of South China Sea Institute of Oceanology, Chinese Academy of Sciences, Guangzhou City, Guangdong Province (http://www.scsio.ac.cn/, Fangfang Yang, [email protected]) under voucher number SY20210603.
3. Methods
Whole-genome DNA was extracted according to a modified CTAB protocol (Doyle Citation1987). The lysis incubation was changed from 30 min to 60 min and then 2 μL RNase A was added at 37 °C for 30 min. The purified genomic DNA was sheared into 350 bp fragments to construct a paired-end (PE) library according to the Nextera XT sample preparation procedures (Illumina, San Diego, CA). The PE reads of 150 bp were generated using a Novaseq 6000 sequencer (Illumina, San Diego, CA) (Fig. S1). A total of 4.15 G of raw data was obtained for further analysis. The GC content, Q20 value and Q30 value of the clean data were 54.94%, 96.53%, and 91.09%, respectively. High-quality reads were assembled into the chloroplast genome using the de novo assembler SPAdes v.3.14.1 software. Finally, the PGA program was used to annotate the chloroplast genome (Qu et al. Citation2019), using the chloroplast genome of Tetraselmis sp. CCMP 881 (GenBank accession number KU167097.1) as the reference. In order to identify the phylogenetic relationship of T. marina, the 12 common protein-coding genes in each complete mitochondrial genome of 22 chloroplast genomes of related marine microalga species were aligned using the MAFFT version 7 software with the FFT-NS-2 strategy (Katoh and Standley Citation2016). Then, a phylogenetic tree was conducted based on the maximum-likelihood method using 1000 bootstrap replicates by IQ-TREE 2.0 (Nguyen et al. Citation2015). Oltmannsiellopsis viridis (GenBank accession number DQ291132.1) was used as an outgroup species.
4. Results
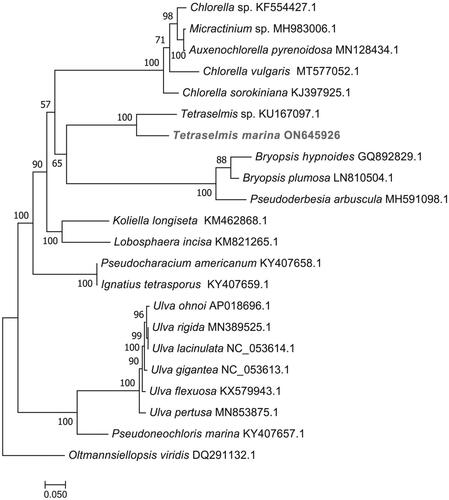
The complete chloroplast genome sequence of T. marina was submitted to GenBank under accession number ON645926. The length of chloroplast genome sequence of T. marina was 96,151 bp, consisting of two inverted repeat regions of 16,158 bp, separated by a large single-copy region of 62,574 bp, and a small single-copy region of 1261 bp (). The overall GC content was 36.6%. There is a cis-splicing gene called atpB (Figure S2). A total of 125 genes were annotated, consisting of 81 protein-coding genes, six rRNA, and 38 tRNA genes. Phylogenetic analysis results showed that T. marina was closely related to Tetraselmis sp. CCMP 881 ().
Figure 2. Gene map of the complete chloroplast genome of T. marina using OGDRAW (https://chlorobox.mpimp-golm.mpg.de/OGDraw.html). Genes shown on the outside of the circle are transcribed clockwise, while those inside are transcribed counterclockwise. Arrangement of 125 genes represented in the map, including 81 protein-coding genes, six rRNA, and 38 tRNA genes.
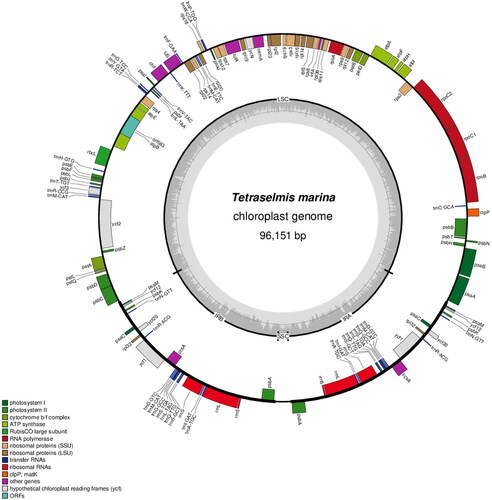
Figure 3. Maximum-likelihood (ML) phylogenetic tree based on complete chloroplast genomes. Oltmannsiellopsis viridis was used as an outgroup species. The numbers on each branches indicate the boot support value of the ML analyses. The scale bar was 0.050. The following sequences were used: Chlorella sp. KF554427.1, Micractinium sp. MH983006.1, Auxenochlorella pyrenoidosa MN128434.1, Chlorella vulgaris MT577052.1, Chlorella sorokiniana KJ397925.1, Tetraselmis sp. CCMP 881 KU167097.1 (Turmel et al. Citation2020), Bryopsis hypnoides GQ892829.1, Bryopsis plumosa GenBank: LN810504.1, Pseudoderbesia arbuscula MH591098.1, Koliella longiseta KM462868.1, Lobosphaera incisa KM821265.1, Pseudocharacium americanum KY407658.1, Ignatius tetrasporus KY407659.1, Ulva ohnoi AP018696.1, Ulva rigida MN389525.1, Ulva lacinulata NC_053614.1, Ulva gigantea NC_053613.1, Ulva flexuosa KX579943.1, Ulva pertusa MN853875.1, Pseudoneochloris marina KY407657.1, and Oltmannsiellopsis viridis DQ291132.1 (Pombert et al. Citation2006).
5. Discussion and conclusions
In this study, the complete chloroplast genome of T. marina was assembled and annotated for the first time. It was 96,151 bp, containing a large single-copy region of 62,574 bp, a small single-copy region of 1,261 bp, and a pair of inverted repeat regions of 16,158 bp. Compared with the chloroplast genome of the marine microalga previously published data, this result indicated that the chloroplast genome of T. marina showed a high level of gene synteny with one publicly available Tetraselmis sp. CCMP 881 (Turmel et al. Citation2020). Phylogenetic trees analysis provided new insight into the genetic relationship of T. marina. Further investigations are necessary to understand and document the evolution of the genus Tetraselmis.
Author contributions
F.F. Yang conceived and wrote the first version of the manuscript; Y. Huang collected the samples and analyzed the data; L.J. Long designed the study. All authors have read and approved the final manuscript.
Ethical approval
The microalga specimen is not designated as endangered species. It requires no specific permissions or licenses. The collection of microalga specimen was legal and reasonable.
Supplemental Material
Download MS Word (1.3 MB)Disclosure statement
The authors declare no potential conflict of interest.
Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI at https://www.ncbi.nlm.nih.gov/ under the accession no. ON645926. The associated BioProject, SRA, and Bio-Sample numbers are PRJNA844566, SRR19521163, and SAMN28834016, respectively.
Additional information
Funding
References
- Doyle JJ. 1987. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull. 19:11–15.
- Fukuda S, Iwamoto K, Atsumi M, Yokoyama A, Nakayama T, Ishida K, Inouye I, Shiraiwa Y. 2014. Global searches for microalgae and aquatic plants that can eliminate radioactive cesium, iodine and strontium from the radio-polluted aquatic environment: a bioremediation strategy. J Plant Res. 127(1):79–89. doi: 10.1007/s10265-013-0596-9.
- Katoh K, Standley DM. 2016. A simple method to control over-alignment in the MAFFT multiple sequence alignment program. Bioinformatics. 32(13):1933–1942. doi: 10.1093/bioinformatics/btw108.
- Martignier A, Filella M, Pollok K, Melkonian M, Bensimon M, Barja F, Langenhorst F, Jaquet JM, Ariztegui D. 2018. Marine and freshwater micropearls: biomineralization producing strontium-rich amorphous calcium carbonate inclusions is widespread in the genus Tetraselmis (Chlorophyta). Biogeosciences. 15(21):6591–6605. doi: 10.5194/bg-15-6591-2018.
- Martignier A, Respinis SD, Filella M, Segovia-Campos I, Marin B, Günther G, Barja F, Tonolla M, Jaquet JM, Melkonian M, et al. 2020. Biomineralization capacities of Chlorodendrophyceae: correlation between chloroplast morphology and the distribution of micropearls in the cell. Protist. 171(5):125760. doi: 10.1016/j.protis.2020.125760.
- Moussa IDB, Chtourou H, Karray F, Sayadi S, Dhouib A. 2017. Nitrogen or phosphorus repletion strategies for enhancing lipid or carotenoid production from Tetraselmis marina. Bioresour Technol. 238:325–332. doi: 10.1016/j.biortech.2017.04.008.
- Nguyen LT, Schmidt HA, Von Haeseler A, Minh BQ. 2015. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 32(1):268–274. doi: 10.1093/molbev/msu300.
- Oliveira Moser GA, Barrera-Alba JJ, Ortega MJ, Alves-de-Souza C, Bartual A. 2022. Comparative characterization of three Tetraselmis chui (Chlorophyta) strains as sources of nutraceuticals. J Appl Phycol. 34(2):821–835. doi: 10.1007/s10811-021-02675-x.
- Pombert JF, Lemieux C, Turmel M. 2006. The complete chloroplast DNA sequence of the green alga Oltmannsiellopsis viridis reveals a distinctive quadripartite architecture in the chloroplast genome of early diverging ulvophytes. BMC Biol. 4(1):3. doi: 10.1186/1741-7007-4-3.
- Qu XJ, Moore MJ, Li DZ, Yi TS. 2019. PGA: a software package for rapid, accurate, and flexible batch annotation of plastomes. Plant Methods. 15(1):50. doi: 10.1186/s13007-019-0435-7.
- Turmel M, Otis C, Cambiaire JCD, Lemieux C. 2020. Complete mitogenomes of the chlorophyte green algae Scherffelia dubia and Tetraselmis sp. CCMP 881 (Chlorodendrophyceae). Mitochondrial DNA B Resour. 5(1):138–139. doi: 10.1080/23802359.2019.1698349.