Abstract
Symbiochlorum hainanensis Gong et al. (Citation2018). is a unicellular green alga belonging to Ulvophyceae, Chlorophyta, and considered as an important species in coral-algae symbiont areas. In this study, we revealed firstly the mitochondrial genome sequence of the S. hainanensis. This mitochondrial genome was a circular DNA molecule of 59,508 bp, including 24 transfer RNA genes, 3 ribosomal RNA genes, and 31 protein-coding genes. The GC content of the genome was 35.4%. The phylogenetic tree suggested that S. hainandiae was a sister to the OUU clade within the class Ulvophyceae. The mitochondrial genome structure and gene content of S. hainanensis supported that S. hainanensis was a new unidentified green alga in Ulvophyceae.
1. Introduction
Symbiochlorum hainanensis Gong et al. (Citation2018), a unicellular green alga, belongs to Ulvophyceae, Chlorophyta, and was firstly identified and described as a new species by Gong et al. (Citation2018). This species is found in various corals in the tropical coral reef areas of the South China Sea. S. hainanensis is a thermo-tolerant alga, which may play vital roles in coral reefs, especially coral-algae symbiont (Gong et al. Citation2020). We also find similar phenomenon that S. hainanensis can maintain rapid growth at high temperature. Mitochondrial genome is known to play important roles in the studies of population genetics, phylogeny, and evolution. In the present study, we analyzed the complete mitochondrial genome of S. hainanensis, and determined its phylogenetic position.
2. Materials
S. hainanensis was collected from coral P. damicornis in the coast of Sanya City, China (18°21′ N and 109°47′ E), and deposited at the laboratory of South China Sea Institute of Oceanology, Chinese Academy of Sciences (Fangfang Yang, [email protected]) under the voucher number SY20210606 ().
3. Methods
The genomic DNA was extracted using the CTAB protocol (Doyle Citation1987). A DNA library was prepared using TruSeq Nano DNA Kit and sequenced on Illumina Novaseq 6000 Sequencing platform (Illumina, CA, USA). The genomic DNA was generated about 4.75 G raw reads, and 4.69 G clean data was obtained from the raw data after quality control processing. The circularized genome was de novo assembled from the high-quality paired-end reads by using the SPAdes 3.14.1 program based on the K-mer size of 105 (Bankevich et al. Citation2012). The mitochondrial genomes were annotated using MFannot and Mitofy (Bernt et al. Citation2013). The average depth was over 200X (Figure S1). The mitochondrial genome map were drawn using the CPGview program (http://www.1kmpg.cn/cpgview/) (Liu et al. Citation2023). Finally, the mitochondrial genome data was uploaded NCBI with accession number ON897766. The 13 common protein-coding genes in each complete mitochondrial genome of 9 species were aligned with the genes in S. hainanensis using MAFFT 7.037 (Katoh and Standley, Citation2016) with the FFT-NS-2 strategy. Then, model-finder var 1.6 was run to select the best-fit model and the cpREV + F + I + G4 model was chosen (Kalyaanamoorthy et al. Citation2017). Finally, iqtree 2.0 was used to construct a phylogenetic tree with 1000 bootstraps based on the ML method (Minh et al. Citation2020).
4. Results
This mitochondrial genome was a circular DNA molecule of 59,508 bp, including 24 transfer RNA genes, 3 ribosomal RNA genes, and 31 protein-coding genes (PCGs) (). The complete mitochondrial genome of S. hainanensis was in the range of known Ulvophyceae mitogenome sizes (55,814–88,416 bp) (Pombert et al. Citation2004; Liu et al. Citation2022). The overall bases were composed of 31.7% A, 32.9% T, 16.6% C, and 18.8% G. Most of the PCGs initiated with the standard ATG except for rps10 ACG with the start codon. Four types of stop codons were TAG (cob, atp8, rpl16, nad1, nad4), TGA (rpl5, tatC rps4, rps13, rps2), GTT (nad7), and the remaining genes with the TAA codon. There are no difficult-to-annotate trans-splicing genes in the mitochondrial genome, but there is a cis-splicing gene called COX1 (Figure S2). Phylogenetic analysis showed that S. hainanensis was sister to OUU clade and TCBD clade within the class Ulvophyceae (Fang et al. Citation2017) ().
Figure 2. The complete mitochondrial genome map of S. hainanensis. Arrangement of 58 genes represented in the map, including 31 protein-coding genes, 24 transfer RNA genes, and 3 ribosomal RNA genes.
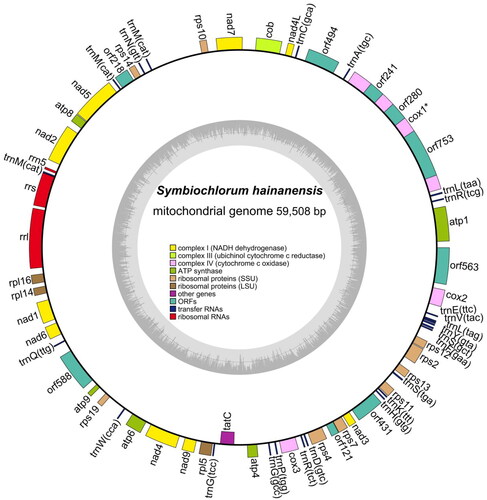
Figure 3. Neighbor-joining phylogenetic tree of S. hainanensis and 8 other closely related species based on the complete mitochondrial genomes. The following sequences were used: Symbiochlorum hainanensis (ON897766), Ulva fenestrata (NC_053629), Ulva ohnoi (AP018695), bryopsis sp. KO-2023 (LC768902), Ulva pertusa (NC_035722), pseudendoclonium akinetum(NC_005926), Ulva pertusa (KX530817), caulerpa lentillifera (KX761577), and monomastix sp. OKE-1(KF060939).

5. Discussion and conclusion
In this study, the complete mitochondrial genome of S. hainanensis was sequenced, assembled, and annotated for the first time. The mitochondrial genome of S. hainanensis was 59,508 bp in length, and it contained 31 protein-coding genes, 24 transfer RNA genes, 3 ribosomal RNA genes. The genomic structure of S. hainanensis was similar to the OUU clade (Zhou et al. Citation2016). This study will enhance the evolution and application study of the class Ulvophyceae.
Ethical approval
The material involved in this paper does not involve endangered or protected species. It requires no specific permissions or licenses. The collection of microalga specimen is legal and reasonable.
Authors’ contribution
Fangfang Yang was involved in the design, acquisition and the drafting of the paper. Lijuan Long was involved in the revision and proving of the paper.
Supplemental Material
Download MS Word (678 KB)Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI at (https://www.ncbi.nlm.nih.gov/) under the accession no. ON897766. The associated BioProject, SRA, and Bio-Sample numbers are PRJNA844566, SRR19521163, and SAMN28834016, respectively.
Additional information
Funding
References
- Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, et al. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 19(5):455–477. doi: 10.1089/cmb.2012.0021.
- Bernt M, Donath A, Juhling F, Externbrink F, Florentz C, Fritzsch G, Putz J, Middendorf M, Stadler PF. 2013. MITOS: improved de novo metazoan mitochondrial genome annotation. Mol Phylogenet Evol. 69(2):313–319. doi: 10.1016/j.ympev.2012.08.023.
- Doyle JJ. 1987. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull. 19:11–15.
- Fang L, Leliaert F, Zhang ZH, Penny D, Zhong BJ. 2017. Evolution of the Chlorophyta: insights from chloro-plast phylogenomic analyses. J Systematics Evol. 55(4):322–332. doi: 10.1111/jse.12248.
- Gong SQ, Jin XJ, Xiao YL, Li ZY. 2020. Ocean acidification and warming lead to increased growth and altered chloroplast morphology in the thermo-tolerant alga Symbiochlorum hainanensis. Front Plant Sci. 11:585202. doi: 10.3389/fpls.2020.585202.
- Gong SQ, Li ZY, Zhang FL, Xiao YL, Cheng H. 2018. Symbiochlorum hainanensis gen. et sp. nov. (Ulvophyceae, Chlorophyta) isolated from bleached corals living in the South China Sea. J Phycol. 54(6):811–817. doi: 10.1111/jpy.12779.
- Katoh K, Standley DM. 2016. A simple method to control over-alignment in the MAFFT multiple sequence alignment program. Bioinformatics. 32(13):1933–1942. doi: 10.1093/bioinformatics/btw108.
- Liu F, Melton JT, III Wang H, Wang J, Lopez-Bautista JM. 2022. Understanding the evolution of mitochondrial genomes in the green macroalgal genus Ulva (Ulvophyceae, Chlorophyta). Front Mar Sci. 9:269. doi: 10.3389/fmars.2022.850710.
- Liu S, Ni Y, Li J, Zhang X, Yang H, Chen H, Liu C. 2023. CPGView: a package for visualizing detailed chloroplast genome structures. Mol Ecol Resour. 23(3):694–704. doi: 10.1111/1755-0998.13729.
- Minh BQ, Schmidt HA, Chernomor O, Schrempf D, Woodhams MD, Haeseler AV, Lanfear R. 2020. IQ-TREE 2: new models and efficient methods for phylogenetic inference in the genomic era. Mol Biol Evol. 37(5):1530–1534. doi: 10.1093/molbev/msaa015.
- Pombert JF, Otis C, Lemieux C, Turmel M. 2004. The complete mitochondrial DNA sequence of the green alga Pseudendoclonium akinetum (Ulvophyceae) highlights distinctive evolutionary trends in the chlorophyta and suggests a sister-group relationship between the Ulvophyceae and Chlorophyceae. Mol Biol Evol. 21(5):922–935. doi: 10.1093/molbev/msh099.
- Kalyaanamoorthy S, Minh BQ, Wong TKF, Haeseler AV, Jermiin LS. 2017. ModelFinder: fast model selection for accurate phylogenetic estimates. Nat Methods. 14(6):587–589. doi: 10.1038/nmeth.4285.
- Zhou L, Wang L, Zhang J, Cai C, He P. 2016. Complete mitochondrial genome of Ulva prolifera, the dominant species of green macroalgal blooms in Yellow Sea, China. Mitochondrial DNA Part B Resour. 1(1):76–78. doi: 10.1080/23802359.2015.1137831.