Abstract
Eirene ceylonensis, a hydrozoan jellyfish species with a complex polymorphic life cycle, is widely distributed in the Chinese coastal sea. In this study, we conducted sequencing and analysis of the first complete mitochondrial genome of E. ceylonensis, obtained from the coastal sea of Qinhuangdao, China. The linear mitochondrial genome is 14,997 bp in length with the overall AT content being 72.8%, encoding 13 protein-coding genes (PCGs), two transfer RNA (tRNA) genes (tRNA-Met and tRNA-Trp) and two ribosomal RNA (rRNA) genes (rrnS and rrnL). Phylogenetic analysis of 13 PCGs suggests that the E. ceylonensis is closely related to Laomedea flexuosa. The availability of the complete mitochondrial genome of E. ceylonensis will be useful for studying the evolutionary relationships of hydrozoan jellyfish species.
1. Introduction
Jellyfish disasters occurred frequently in the coastal water of Qinhuangdao in the Bohai Sea, which is known for its high diversity of jellyfish species (Yuan et al. Citation2021). The hydrozoan jellyfish are a diverse group of aquatic animals with various morphologies and life cycles, and hydrozoan is an ideal model for studying germ cell development due to their simplicity of organization and manipulation, as well as their transparency (Amiel et al. Citation2010). Eirene ceylonensis Browne Citation1905, once named as the Irene ceylonensis, was first collected from the Galle Bay in the Gulf of Manaar (Browne Citation1905). It is a typical marine hydrozoan species in the family Eirenidae (Cnidaria: Hydrozoa: Leptothecata), has a worldwide distribution and is also widely found in the Chinese coastal seas (Zhang Citation1979). In recent years, E. ceylonensis has become a dominant species in many areas of the Bohai Sea, including the southwest part (Liu et al. Citation2023), Qinhuangdao coastal waters (Xu et al. Citation2022), and the Yellow River estuary and its adjacent area (Li et al. Citation2018). Eirene ceylonensis has a complex polymorphic life cycle that includes both the medusa and polyp phases, with the medusa phase having an umbrella diameter of only about 15–25 mm (Xu et al. Citation2014). And the morphology of the jellyfish varies greatly in different periods, so these characteristics may hinder accurate identification of the jellyfish species. Therefore, we conducted mitogenome sequencing of E. ceylonensis. The first complete mitogenome of Eirene species will be useful for species identification, phylogenetic studies, and biogeographical research of hydrozoan jellyfish.
2. Materials and methods
2.1. Sample collection
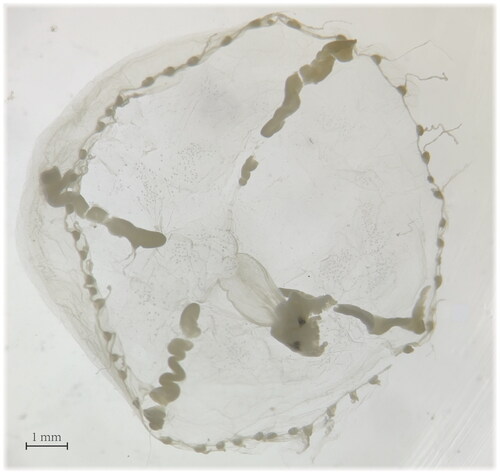
A total of five E. ceylonensis specimens were collected from the coastal sea of Qinhuangdao (39.916°N, 119.620°E) in the Bohai Sea, China, using the plankton net on 13 August 2022. Firstly, the specimens (voucher CMY22Q801-CMY22Q805) underwent microscopic examination for identification purpose prior to genome sequencing. These specimens have the same physiological structure characteristics, except for the umbrella diameter. For example, the voucher CMY22Q801 (), which flat and thin umbrella is about 10 mm in diameter with 48 tentacles, and four linear gonads extending from the base of the peduncle to near the margin of the umbrella. These characteristics are basically consistent with the hydrozoan jellyfish E. ceylonensis (Browne Citation1905). Then, the voucher CMY22Q801 was frozen using liquid nitrogen for the DNA extraction. The remaining specimens were stored in absolute ethanol and deposited at Hebei Normal University of Science and Technology (https://www.hevttc.edu.cn/, contact person: Yang Chen, email: [email protected]).
2.2. Methods
Total DNA was extracted from the muscle tissue of E. ceylonensis voucher CMY22Q801 using a DNeasy Blood & Tissue Kit (QIAGEN, Germany) according to the manufacturer’s instructions. The whole genome was sequenced using an Illumina Hiseq 2500 platform (paired-end 150 bp reads) at Novogene Co., Ltd. (Beijing, China). The sequencing results (about 16.86 M raw paired reads) were assembled into the mitogenome using GetOrganelle v 1.7.2 (Jin et al. Citation2020), with SPAdes v 3.13.2 as the assembler (Bankevich et al. Citation2012). The read coverage depth map (Figure s1), extracted using the Integrative Genomics Viewer (Robinson et al. Citation2011), is presented in supplementary material. The ORFfinder and MITOS (Bernt et al. Citation2013) were used to identify the protein-coding genes (PCGs), transfer RNA (tRNA) genes, and ribosomal RNA (rRNA) genes in the mitogenome. The gene map of E. ceylonensis mitogenome was generated using the Organellar Genome DRAW (Greiner et al. Citation2019).
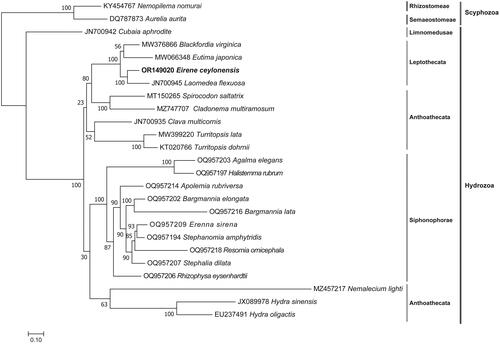
By searching the NT database (www.ncbi.nlm.nih.gov), there were 22 hydrozoan species with complete mitogenome sequences available, including 12 complete mitochondrial genomes with associated publications (). For phylogenetic analysis, the 23 complete mitochondrial genomes from hydrozoan species were included, including E. ceylonensis. To enhance the resolution of the phylogenetic relationship among hydrozoan jellyfish species, two mitogenomes of Scyphozoa species of Cnidaria, Nemopilema nomurai (KY454767) (Wang and Sun Citation2017) and Aurelia aurita (DQ787873) (Shao et al. Citation2006) were used as outgroup. Amino acid sequences of 13 PCGs from the 25 species were individually aligned using MAFFT v7.471 (Katoh and Standley Citation2013). The alignments were then trimmed using trimal v1.2 (Capella-Gutiérrez et al. Citation2009) and concatenated using Phyutility (Smith and Dunn Citation2008). The incongruence length difference (ILD) test was conducted using PAUP4.0 (Swofford Citation2002) with the same parameters in our previous work (Chen et al. Citation2021). Finally, a Maximum-likelihood (ML) phylogenetic tree was constructed using IQtree v1.6.12 (Trifinopoulos et al. Citation2016) with 1000 bootstraps replicates.
Table 1. The 25 complete mitochondrial genomes of Cnidaria species used for phylogenetic analysis in this study.
3. Results
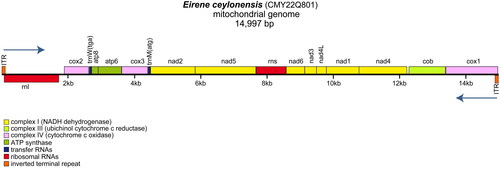
Through microscopic examination, the voucher CMY22Q801 can be basically determined as E. ceylonensis (). The complete mitogenome of E. ceylonensis is linear in structure and 14,997 bp in length (GenBank accession number OR149020), with high AT bias (A: 32.0%; T: 40.8%; C: 12.4%; G: 14.8%). Within the mitogenome of E. ceylonensis, 13 protein-coding genes (cox2, atp8, atp6, cox3, nad2, nad5, nad6, nad3, nad4L, nad1, nad4, cob, cox1), two tRNAs (tRNA-Met and tRNA-Trp), and two rRNAs (rrnS and rrnL) were encoded. Most of these genes were located on the plus strand, except for the rrnL, which resided on the minus strand (). Moreover, an inverted terminal repeat (ITR) sequence with 105 bp in length was found in the mitogenome of E. ceylonensis (Figure s2).
The ILD test (p = 0.01) indicated that amino acid sequence concatenation of 13 PCGs from 25 species would not affect phylogenetic accuracy (Cunningham Citation1997). The ML phylogenetic tree revealed that E. ceylonensis mitogenome formed a cluster with three other Leptothecata species mitogenomes ().
4. Discussion and conclusions
The mitogenome of E. ceylonensis exhibits a linear molecular structure with a length of 14,997 bp, which is comparatively shorter than that of three other Leptothecata species, namely L. flexuosa (16,075 bp) (Kayal et al. Citation2012), Eutima japonica (15,315 bp) (Seo et al. Citation2021a) and Blackfordia virginica (15,109 bp) (Seo et al. Citation2021b). However, the linear molecular structure of the E. ceylonensis mitogenome is consistent with the aforementioned Leptothecata species. High AT bias (A + T: 72.8%) is found in the E. ceylonensis mitogenome, which is similar to other hydrozoan mitogenomes, such as the Nemalecium lighti (A + T: 71.8%) (Macher et al. Citation2021) and B. virginica (A + T: 73.6%) (Seo et al. Citation2021b). The gene map of the E. ceylonensis mitogenome resembles those of Clytia hemisphaerica, particularly the ITR structure in the C. hemisphaerica mitogenome (GenBank accession number CACVBU010001317) (Leclère et al. Citation2019). Repeat sequences have been discovered in many hydrozoan mitochondrial genomes with different length, such as the Hydra oligactis and Liriope tetraphylla (Kayal et al. Citation2015). However, the function of these ITR structures is still unclear and deserves further exploration. The first reported mitogenome sequence of E. ceylonensis in this study will enhance our understanding of the phylogenetic relationship of Eirene species. Moreover, it will contribute to future research in phylogenetic analysis, population genetics, and biogeography of the hydrozoans.
Ethical approval
The jellyfish species is not included in the 'List of Protected Animals in China’, and the study did not involve endangered or protected species. The approval of sample collection is not required according to the Animal Ethical and Welfare Committee of Hebei Normal University of science and Technology.
Author contribution
YC, conceived and designed the experiments, analyzed the data, prepared figures, and approved the final draft. SD, analyzed the data, prepared figures, and approved the final draft. DS, analyzed the data, prepared figures, and approved the final draft. YW, analyzed the data, prepared figures, reviewed drafts of the paper. JN, analyzed the data, prepared figures, reviewed drafts of the paper. ZL, conceived and designed the experiments, reviewed drafts of the paper.
Supplemental Material
Download JPEG Image (404 KB)Supplemental Material
Download TIFF Image (26.7 MB)Acknowledgement
We would like to thank Dr Xiaobo Yuan for his help in fieldwork and sampling.
Disclosure statement
No potential conflict of interest was reported by the authors.
Data availability statement
The genome sequence data supporting this study’s findings are openly available in GenBank of NCBI at https://www.ncbi.nlm.nih.gov/under the accession number OR149020. The associated BioProject, Bio-Sample and SRA numbers are PRJNA985931, SAMN35816308, and SRR24984782, respectively.
Additional information
Funding
References
- Amiel A, Chang P, Momose T, Houliston E. 2010. Clytia hemisphaerica: a Cnidarian model for studying oogenesis. West Sussex, UK: John Wiley & Sons Ltd, p. 501.
- Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, et al. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 19(5):455–477. doi:10.1089/cmb.2012.0021.
- Bernt M, Donath A, Jühling F, Externbrink F, Florentz C, Fritzsch G, Pütz J, Middendorf M, Stadler PF. 2013. MITOS: Improved de novo metazoan mitochondrial genome annotation. Mol Phylogenet Evol. 69(2):313–319. doi:10.1016/j.ympev.2012.08.023.
- Browne ET. 1905. Report on the medusae (Hydromedusae, Scyphomedusae and Ctenophora) collected by Prof. Herdman at Ceylon in 1902. Government of Ceylon on Pearl Oyster fisheries of the Gulf of Manaar. London, UK: University College London. 4:132–166.
- Capella-Gutiérrez S, Silla-Martínez JM, Gabaldón T. 2009. trimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics. 25(15):1972–1973. doi:10.1093/bioinformatics/btp348.
- Chen Y, Wang Y, Liu K, Liu F, Chen N. 2021. Development of a high-resolution molecular marker for tracking Pseudo-nitzschia pungens genetic diversity through comparative analysis of mitochondrial genomes. J Appl Phycol. 33(4):2283–2298. doi:10.1007/s10811-021-02461-9.
- Cunningham CW. 1997. Can three incongruence tests predict when data should be combined? Mol Biol Evol. 14(7):733–740. doi:10.1093/oxfordjournals.molbev.a025813.
- Fang X, Zhou K, Chen J. 2022. The complete linear mitochondrial genome of the hydrozoan jellyfish Cladonema multiramosum Zhou et al., 2022(Cnidaria: Hydrozoa: Cladonematidae). Mitochondrial DNA Part B Resour. 7(6):921–923. doi:10.1080/23802359.2022.2079100.
- Greiner S, Lehwark P, Bock R. 2019. OrganellarGenomeDRAW (OGDRAW) version 1.3.1: expanded toolkit for the graphical visualization of organellar genomes. Nucleic Acids Res. 47(W1):W59–W64. doi:10.1093/nar/gkz238.
- Jin J-J, Yu W-B, Yang J-B, Song Y, dePamphilis CW, Yi T-S, Li D-Z. 2020. GetOrganelle: a fast and versatile toolkit for accurate de novo assembly of organelle genomes. Genome Biol. 21(1):241. doi:10.1186/s13059-020-02154-5.
- Katoh K, Standley DM. 2013. MAFFT Multiple sequence alignment Software Version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780. doi:10.1093/molbev/mst010.
- Kayal E, Lavrov DV. 2008. The mitochondrial genome of Hydra oligactis (Cnidaria, Hydrozoa) sheds new light on animal mtDNA evolution and cnidarian phylogeny. Gene. 410(1):177–186. doi:10.1016/j.gene.2007.12.002.
- Kayal E, Bentlage B, Collins AG, Kayal M, Pirro S, Lavrov DV. 2012. Evolution of linear mitochondrial genomes in Medusozoan Cnidarians. Genome Biol Evol. 4(1):1–12. doi:10.1093/gbe/evr123.
- Kayal E, Bentlage B, Cartwright P, Yanagihara AA, Lindsay DJ, Hopcroft RR, Collins AG. 2015. Phylogenetic analysis of higher-level relationships within Hydroidolina (Cnidaria: Hydrozoa) using mitochondrial genome data and insight into their mitochondrial transcription. PeerJ. 3:e1403. doi:10.7717/peerj.1403.
- Leclère L, Horin C, Chevalier S, Lapébie P, Dru P, Peron S, Jager M, Condamine T, Pottin K, Romano S, et al. 2019. The genome of the jellyfish Clytia hemisphaerica and the evolution of the cnidarian life-cycle. Nat Ecol Evol. 3(5):801–810. doi:10.1038/s41559-019-0833-2.
- Li H, Liu G, Ma J, Chen H. 2018. Community characteristics of zooplankton in the Yellow River estuary and its adjacent area in summer and autumn. Mar Environ Sci. 37(5):631–639. doi:10.13634/j.cnki.mes.2018.05.002.
- Lisenkova AA, Grigorenko AP, Tyazhelova TV, Andreeva TV, Gusev FE, Manakhov AD, Goltsov AY, Piraino S, Miglietta MP, Rogaev EI. 2017. Complete mitochondrial genome and evolutionary analysis of Turritopsis dohrnii, the "immortal" jellyfish with a reversible life-cycle. Mol Phylogenet Evol. 107:232–238. doi:10.1016/j.ympev.2016.11.007.
- Liu X, Xu Z, Zhang H, Li B, Wen G, Li X, Sha J, Du X, Bao M, Sun Z, et al. 2023. Characteristics of small medusae community correlated with environmental factors in the southwest of Bohai Sea. Haiyang Xuebao. 45(3):66–75. doi:10.12284/hyxb2023012.
- Macher J-N, Kayal E, Duijm E, van der Hoorn B, Montano S, Speksnijder A. 2021. The mitochondrial genome of Nemalecium lighti (Hydrozoa, Leptothecata). Mitochondrial DNA Part B Resour. 6(11):3196–3198. doi:10.1080/23802359.2021.1989335.
- Pan HC, Qian XC, Li P, Li XF, Wang AT. 2014. The complete mitochondrial genome of Chinese green hydra, Hydra sinensis (Hydroida: Hydridae). Mitochondrial DNA. 25(1):44–45. doi:10.3109/19401736.2013.782017.
- Robinson JT, Thorvaldsdóttir H, Winckler W, Guttman M, Lander ES, Getz G, Mesirov JP. 2011. Integrative genomics viewer. Nat Biotechnol. 29(1):24–26. doi:10.1038/nbt.1754.
- Seo Y, Chae J, Ki J-S. 2020. The complete mitochondrial genome of the hydrozoan jellyfish Spirocodon saltatrix (Cnidaria; Hydrozoa; Anthoathecata) with phylogeny analysis. Mitochondrial DNA Part B Resour. 5(3):3116–3117. doi:10.1080/23802359.2020.1797568.
- Seo JS, Eom H-J, Cho J-K, Kang H-S, Rhee J-S. 2021a. The linear mitochondrial genome of commensal hydroid Eutima japonica (Cnidaria, Hydrozoa, Eirenidae). Mitochondrial DNA Part B Resour. 6(3):1082–1084. doi:10.1080/23802359.2021.1899869.
- Seo Y, Chae J, Ki J-S. 2021b. Complete mitochondrial genome of the hydrozoan jellyfish Blackfordia virginica Mayer, 1910 (Cnidaria; Hydrozoa; Leptothecata) with phylogenetic analysis. Mitochondrial DNA Part B Resour. 6(3):1202–1203. doi:10.1080/23802359.2021.1903363.
- Seo Y, Chae J, Ki J-S. 2021c. The complete mitochondrial genome of the hydrozoan jellyfish Turritopsis lata Lendenfeld, 1885 (Cnidaria; Hydrozoa; Anthoathecata) with molecular phylogenetic analysis. Mitochondrial DNA Part B Resour. 6(7):1992–1993. doi:10.1080/23802359.2021.1938725.
- Shao Z, Graf S, Chaga OY, Lavrov DV. 2006. Mitochondrial genome of the moon jelly Aurelia aurita (Cnidaria, Scyphozoa): a linear DNA molecule encoding a putative DNA-dependent DNA polymerase. Gene. 381:92–101. doi:10.1016/j.gene.2006.06.021.
- Smith SA, Dunn CW. 2008. Phyutility: a phyloinformatics tool for trees, alignments and molecular data. Bioinformatics. 24(5):715–716. doi:10.1093/bioinformatics/btm619.
- Swofford DL. 2002. PAUP*: Phylogenetic Analysis Using Parsimony. Sunderland, MA: Sinauer Associates.
- Trifinopoulos J, Nguyen L-T, von Haeseler A, Minh BQ. 2016. W-IQ-TREE: a fast online phylogenetic tool for maximum likelihood analysis. Nucleic Acids Res. 44(W1):W232–W235. doi:10.1093/nar/gkw256.
- Wang Y, Sun S. 2017. Complete mitochondrial genome of the jellyfish, Rhopilema esculentum Kishinouye 1891 (Cnidaria: Scyphozoa) and the phylogenetic relationship in the related species. Mitochondrial DNA Part B Resour. 2(1):167–168. doi:10.1080/23802359.2017.1303342.
- Xu ZZ, Huang JQ, Lin M, Guo DH, Wang CG. 2014. The superclass Hydrozoa of the phylum Cnidaria in China. Beijing, China: J Ocean Press. p. 519.
- Xu D, Qi Y, Zhang X, Du X, Han L, Liu X, Zhou H, Pan Y. 2022. Distribution characteristics of small medusae in the Qinhuangdao coastal waters during summer. Marine Sciences. 46(11):29–37. doi:10.11759/hykx20211013001.
- Yuan X, Liu Z, Xue L, Chen X, An Y. 2021. Review on the occurrence regularity and current situation of jellyfish disaster in Qinhuangdao coastal area. Hebei Fisheries. 6:12–17. doi:10.3969/j.issn.1004-6755.2021.06.004.
- Zhang J. 1979. A preliminary analysis on the hydromedusae fauna of the China sea areas. Acta Oceanolog Sin. 1(1):127–137.