Abstract
In this study, the complete mitogenome of the entomopathogenic fungus Metarhizium pinghaense 15 R, which is highly virulent to aphids and was isolated from Korean soil, was assembled and annotated for three ATP synthase subunits (atp6, atp8, and atp9), three cytochrome oxidase subunits (cox1, cox2, and cox3), apocytochrome b (cob), seven subunits of NADH dehydrogenase (nad1, nad2, nad3, nad4, nad4L, nad5, and nad6), two ribosomal RNAs (rnl and rns), and 19 tRNA genes. Five genes were carrying a total of eight introns, and they may encode ribosomal protein S3, LAGLIDADG and GIY-YIG endonucleases. Phylogenetic analysis based on the mitochondrial nucleotide sequence confirmed that the M. pinghaense 15 R is a member of the Clavicipitaceae, and is closely related to the species M. anisopliae, M. robertsii, and M. brunneum. The mtDNA base sequence of the M. pinghaense 15 R strain reported in this study is thought to be useful for biological resource genetic data.
Introduction
Metarhizium pinghaense Q.T. Chen & H.L. Guo, 2020 is a fungus that causes disease in insects and belongs to the family Clavicipitaceae (Ascomycota: Hypocreales). The fungal genus Metarhizium, characterized by the formation of green conidia after killing a host insect, was first described by Metschnikoff in 1879 (Zimmermann et al. Citation1995), and has been isolated from various soils around the world, making it one of the most famous entomopathogenic fungi to date (Shin et al. Citation2013). Among species belonging to the genus Metarhizium, the fungus belongs to the most recently differentiated group, with a wide host range from endopterygota to exopterygota of the insect class, and even spider class (Mongkolsamrit et al. Citation2020). The genus Metarhizim including this species is recognized as an important entomopathogenic fungal group for coevolution studies with insects through genome analysis (Hu et al. Citation2014). As a result of these interests, the whole genome and mitochondrial sequence of this fungal species have already been determined and used in various studies (Ghikas et al. Citation2006; Gao et al. Citation2011). To our knowledge, the complete mitochondrial sequence of M. brunnem, M. anisoplie, M. album, and M. rileyi was released in NCBI database (National Center for Biotechnology Information, https://www.ncbi.nlm.nih.gov/). However, the complete mitochondrial sequence of M. pinghaense is still unreported. In addition, to secure the population genetics and biological sovereignty for use as microbial pesticides of this worldwide fungal species, it is necessary to secure the mitochondrial sequence of the fungal species isolated from various regions. Therefore, in this study, we present the complete mitogenome of the M. pinghaense 15 R strain isolated in Korea, which shows high aphid-insecticidal activity (Heo et al. Citation2023). The M. pinghaense 15 R strain is molecularly characterized by the findings of this study, which can also be used to classify and investigate the evolution of the entomopathogenic fungi Metarhizuum.
Materials and methods
The M. pinghaense 15 R strain was isolated from a Korean soil sample (36°37'43.2"N 127°27'08.1"E), and deposited in the Korean Agricultural Culture Collection (http://genebank.rda.go.kr/, ByeongHak Han, [email protected] ) under the voucher number KACC 83065BP (). This strain shows strong pathogenicity against aphids and is receiving positive attention in research on the development of microbial pesticides.
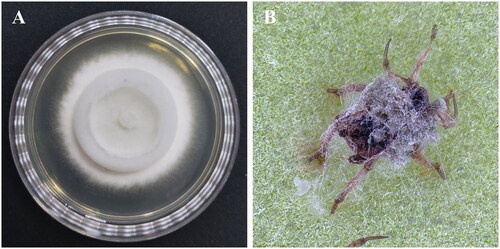
Figure 1. Morphological observation of the Metarhizium pinghaense 15 R strain. (A), the strain was cultured on PDA medium at 25 °C for 2 weeks; (B), green peach aphid (Myzus persicae) infected with the strain. The cadaver was covered with the strain’s white hyphae and green conidia and was photographed by InJi Heo at the insect pathology and bioactives laboratory of jeonbuk National university, Korea, on 19 jan 2023.
The total DNA of M. pinghaense 15 R was fragmented by sonication to a size of ∼280 bp, followed by sequencing on an Illumina NovaSeq platform in 2 × 150 bp reads. The high quality PE reads were assembled by CLC assembly cell (ver. 4.010.83648, CLC QIAGEN) and Mitoz (Meng et al. Citation2019), followed by manual curation through PE reads mapping (Kim et al. Citation2015). Annotation of the complete mitochondrial genome was performed with Geseq (Tillich et al. Citation2017) and manual corrections (Meng et al. Citation2019). The coverage map was visualized with the tablet program (Milne et al. Citation2010).
For the phylogenetic relationship of Metarhizium pinghaense 15 R strain, the phylogenetic tree was constructed based on the complete genomes of 11 species with Verticillium dahliae NC_008248.1 as the outgroup. Multiple sequence alignments were performed using the MAFFT 7.520 program (Rozewicki et al. Citation2019), and phylogenetic analysis was performed Maximum likelihood (ML) method of RAxML 8.2.12 (Stamatakis Citation2014) using the nucleotide substitution model GTR + G + I model with 1,000 bootstraps replicates. The following sequences with GenBank accession were used: M. album MW448543.1 (Sun et al. Citation2021), M. anisopliae NC_008068.1 (Ghikas et al. Citation2006), M. brunneum LR792747.1 (Kortsinoglou et al. Citation2020), M. rileyi MT107156.1 (Zhang et al. Citation2020a), M. robertsii JELW01000367, Akanthomyces lecanii MN904747.1 (Zhang et al. Citation2020b), Clonostachys rosea NC_036667.1, Fusarium solani NC_016680.1 (Al-Reedy et al. Citation2012), Gibberella moniliformis NC_016687.1 (Al-Reedy et al. Citation2012), Hirsutella thompsonii MH367296.1 (Wang et al. Citation2018), Metacordyceps chlamydosporia NC_022835.1 (Lin et al. Citation2015), and Verticillium dahliae NC_008248.1 (Pantou et al. Citation2006).
Results
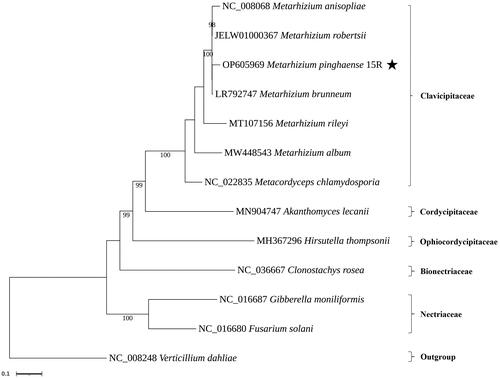
The mitogenome of M. pinghaense 15 R (GenBank accession: OP605969.1) was a circular molecule of 35,763 bp with GC content of 27% (). The average and maximum coverage were 3654.8x and 6445x, respectively (Figure S1). Identified genes include, the three ATP synthase subunits 6, 8, and 9 (atp6, atp8, and atp9), the three cytochrome oxidase subunits I, II, III (cox1, cox2, and cox3) and apocytochrome b (cob), the seven subunits of NADH dehydrogenase (nad1, nad2, nad3, nad4, nad4L, nad5, and nad6), the two ribosomal RNAs (rnl and rns), and 19 tRNA genes. Five genes were carrying a total of eight introns, including cox1, cob, and rnl with two introns while cox3 and nad1 with one intron each. These introns all belonged to the group I intron family, and they may encode ribosomal protein S3 (in rnl), LAGLIDADG (in cob, cox1, and cox3), and GIY-YIG endonucleases (in cob). The structures of the cis-splicing genes are shown in Figure S2. Phylogenetic analysis based on the mitochondrial nucleotide sequence (six conserved protein-coding sequences of cob, cox1, cox2, nad1, nad4, and nad5) by the Maximum Likelihood approach identified the M. pinghaense 15 R as a member of the Clavicipitaceae ().
Figure 2. Graphic representation of19 features identified the Metarhizium pinghaense 15 R mitochondrial genome. The map was prepared using OGDRAW program (https://chlorobox.mpimp-golm.mpg.de/OGDraw.html). Genes are shown outside, and GC and at contents across the mitochondrial genome are shown with dark and light shading, respectively, inside the inner circle. Genes are color-coded by their functional classification.
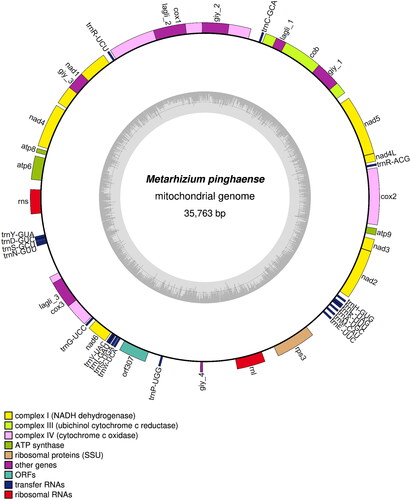
Figure 3. Phylogenetic trees based on the mitochondrial nucleotide sequence in related fungal species. Phylogenetic trees based on the mitochondrial nucleotide sequence in related fungal species. Phylogenetic tree of mitochondrial genomes of Metarhizium pinghaense 15 R and its related 12 species. We used seven species of Clavicipitaceae and representative species of other families with available mitogenomes in Hypocreales. Protein-coding sequences conserved in the mitochondrial genomes of 13 species were multiple-aligned using MAFFT (http://mafft.cbrc.jp/alignment/server/index.html) and used to generate a phylogenetic tree using the maximum likelihood approach as implemented in RAxML v8.2.12 (StamatakisCitation2014). the bootstrap values were indicated for nodes that support values of >70%. and Verticillium dahliae NC_008248.1 (Pantou et al. Citation2006) used an outgroup of this tree. The species under study is highlighted by the star. The following sequences were used: Metarhizium album MW448543.1 (Sun et al. Citation2021), Metarhizium anisopliae NC_008068.1 (Ghikas et al. Citation2006), Metarhizium brunneum LR792747.1 (Kortsinoglou et al. Citation2020), Metarhizium rileyi MT107156.1 (Zhang et al. Citation2020a), Metarhizium robertsii JELW01000367, Akanthomyces lecanii MN904747.1 (Zhang et al. Citation2020b), Clonostachys rosea NC_036667.1, Fusarium solani NC_016680.1 (Al-Reedy et al. Citation2012), Gibberella moniliformis NC_016687.1 (Al-Reedy et al. Citation2012), Hirsutella thompsonii MH367296.1 (Wang et al. Citation2018), Metacordyceps chlamydosporia NC_022835.1 (Lin et al. Citation2015), and Verticillium dahliae NC_008248.1 (Pantou et al. Citation2006).
Discussion and conclusion
This study was conducted as part of the work to characterize the M. pinghaense 15 R strain. The genus Metarhizium, the subject of this study, is one of the pathogenic fungi that cause insect diseases. Furthermore, these fungi have been produced and used as an environmentally friendly pest control method (Sullivan et al. Citation2022). At the same time as the preference for eco-friendly control measures is increasing, more microbial pesticides are being developed compared to the past, and research is also needed to secure the sovereignty of biological resources of strains developed as microbial pesticides (Lee et al. Citation2018). In our knowledge, current efforts to secure the sovereignty of these microorganisms include whole genome analysis, but there is no approach using mtDNA. Since it takes a lot of time and effort to analyze the whole genome (ca. 30 ∼ 41 Mb), which is relatively large in size, it is considered more accessible to secure the sovereignty of biological resources by securing the mtDNA sequence (ca. 24 ∼ 68 Kb). In other words, it is thought that the mtDNA sequence of the M. pinghaense 15 R strain reported in this study can be helpful as genetic data for biological resources.
So far, three species (M. anisoplie NC_008068.1, M. brunnem LR792747.1, and M. robertsii JELW01000367) of the Metarhizium have been analyzed for the main purpose of securing the mtDNA sequence, and only the mtDNA sequence of the other two species (M. album MW448543.1 and M. rileyi MT107156.1) has been determined through whole genome analysis. Of the approximately 55 species recorded in this genus, only 12% have been analyzed for mtDNA (Bischoff et al. Citation2009; Lopes et al. Citation2018; Mongkolsamrit et al. Citation2020;). This study is the first report analyzing and reporting the Metarhizium pinghaense mtDNA sequence to the best of our knowledge.
The number of tRNAs revealed the difference between M. pinghaense and M. anisopliae, despite their tight evolutionary relationship. In a previous study, M. anisopliae had 24 tRNAs (Ghikas et al. Citation2006), whereas M. pinghaense 15 R had 19 tRNAs. In comparing tRNA types, M. pinghaense 15 R have proline and valine, which were not confirmed in M. anisopliae of tRNA. In contrast, glutamine, phenylalanine, and threonine were not confirmed in M. pinghaense 15 R strain.
As a result, this fungus showed a close relationship with the existing M. anisopliae and M. robertsii of the same genus, which supports the recent differentiation of M. anisopliae and M. guizhouense (Mongkolsamrit et al. Citation2020). The M. pinghaense mtDNA sequence obtained through this study will be helpful for taxonomic and evolution studies as important data to which this fungus belongs.
Ethical approval
No ethical approval was required for this study.
Authors’ contributions
All authors contributed to the study conception and design; Material preparation, data collection and analysis were performed by Tae Young Shin; Seulki Kim and InJi Heo were involved in the acquisition, analysis and interpretation of the data; Soo Dong Woo critically reviewed the article; The first draft of the manuscript was written by Woo Jin Kim. All authors commented on previous versions of the manuscript. All authors read and approved the final manuscript.
Supplemental Material
Download MS Word (94 KB)Supplemental Material
Download MS Word (68.3 KB)Disclosure statement
No potential conflict of interest was reported by the authors.
Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI at https://www.ncbi.nlm.nih.gov/ under the accession no. OP605969. The associated BioProject, SRA, and Bio-Sample numbers are PRJNA815910, SRR18320042, and SAMN26650411, respectively.
Additional information
Funding
References
- Al-Reedy RM, Malireddy R, Dillman CB, Kennell JC. 2012. Comparative analysis of Fusarium mitochondrial genomes reveals a highly variable region that encodes an exceptionally large open reading frame. Fungal Genet Biol. 49(1):2–14. doi: 10.1016/j.fgb.2011.11.008.
- Bischoff JF, Rehner SA, Humber RA. 2009. A multilocus phylogeny of the Metarhizium anisopliae lineage. Mycologia. 101(4):512–530. doi: 10.3852/07-202.
- Gao Q, Jin K, Ying S-H, Zhang Y, Xiao G, Shang Y, Duan Z, Hu X, Xie X-Q, Zhou G, et al. 2011. Genome sequencing and comparative transcriptomics of the model entomopathogenic fungi Metarhizium anisopliae and M. acridum. PLOS Genet. 7(1):e1001264. doi: 10.1371/journal.pgen.1001264.
- Ghikas DV, Kouvelis VN, Typas MA. 2006. The complete mitochondrial genome of the entomopathogenic fungus Metarhizium anisopliae var. anisopliae: gene order and trn gene clusters reveal a common evolutionary course for all Sordariomycetes, while intergenic regions show variation. Arch Microbiol. 185(5):393–401. doi: 10.1007/s00203-006-0104-x.
- Heo I, Kim S, Han GH, Im S, Kim JW, Hwang DY, Jang JW, Lee JY, Woo SD, Shin TY. 2023. Characteristics of insecticidal substances from the entomopathogenic fungus Metarhizium pinghaense 15R against cotton aphid in Korea. J Asia-Pac Entomol. 26(1):102013. doi: 10.1016/j.aspen.2022.102013.
- Hu X, Xiao G, Zheng P, Shang Y, Su Y, Zhang X, Liu X, Zhan S, Leger RJS, Wang C. 2014. Trajectory and genomic determinants of fungal-pathogen speciation and host adaptation. Proc Natl Acad Sci U S A. 111(47):16796–16801. doi: 10.1073/pnas.1412662111.
- Rozewicki J, Li S, Amada KD, Standley DM, Katoh K. 2019. MAFFT-DASH: integrated protein sequence and structural alignment (describes web interface for sequence and structural alignments). Nucleic Acids Res. 47(W1):W5–W10. doi: 10.1093/nar/gkz342.
- Kim K, Lee S-C, Lee J, Yu Y, Yang K, Choi B-S, Koh H-J, Waminal NE, Choi H-I, Kim N-H, et al. 2015. Complete chloroplast and ribosomal sequences for 30 accessions elucidate evolution of Oryza AA genome species. Sci Rep. 5(1):15655. doi: 10.1038/srep15655.
- Kortsinoglou AM, Saud Z, Eastwood DC, Butt TM, Kouvelis VN. 2020. The mitochondrial genome contribution to the phylogeny and identification of Metarhizium species and strains. Fungal Biol. 124(10):845–853. doi: 10.1016/j.funbio.2020.06.003.
- Lee SJ, Lee MR, Kim S, Kim JC, Park SE, Li D, Shin TY, Nai Y, Kim JS. 2018. Genomic analysis of the insect-killing fungus Beauveria bassiana JEF-007 as a biopesticide. Sci Rep. 8(1):12388. doi: 10.1038/s41598-018-30856-1.
- Lin R, Liu C, Shen B, Bai M, Ling J, Chen G, Mao Z, Cheng X, Xie B. 2015. Analysis of the complete mitochondrial genome of Pochonia chlamydosporia suggests a close relationship to the invertebrate-pathogenic fungi in Hypocreales. BMC Microbiol. 15(1):5. doi: 10.1186/s12866-015-0341-8.
- Lopes RB, Souza DA, Rocha LF, Montalva C, Luz C, Humber RA, Faria M. 2018. Metarhizium alvesii sp. nov.: a new member of the Metarhizium anisopliae species complex. J Invertebr Pathol. 151:165–168. doi: 10.1016/j.jip.2017.12.001.
- Meng G, Li Y, Yang C, Liu S. 2019. MitoZ: a toolkit for animal mitochondrial genome assembly, annotation and visualization. Nucleic Acids Res. 47(11):e63-e63–e63. doi: 10.1093/nar/gkz173.
- Milne I, Bayer M, Cardle L, Shaw P, Stephen G, Wright F, Marshall D. 2010. Tablet—next generation sequence assembly visualization. Bioinformatics. 26(3):401–402. doi: 10.1093/bioinformatics/btp666.
- Mongkolsamrit S, Khonsanit A, Thanakitpipattana D, Tasanathai K, Noisripoom W, Lamlertthon S, Himaman W, Houbraken J, Samson R, Luangsa-Ard J. 2020. Revisiting Metarhizium and the description of new species from Thailand. Stud Mycol. 95(1):171–251. doi: 10.1016/j.simyco.2020.04.001.
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30(9):1312–1313. doi: 10.1093/bioinformatics/btu033.
- Pantou MP, Kouvelis VN, Typas MA. 2006. The complete mitochondrial genome of the vascular wilt fungus Verticillium dahliae: a novel gene order for Verticillium and a diagnostic tool for species identification. Curr Genet. 50(2):125–136. doi: 10.1007/s00294-006-0079-9.
- Shin TY, Lee WW, Ko SH, Choi JB, Bae SM, Choi JY, Lee KS, Je YH, Jin BR, Woo SD. 2013. Distribution and characterisation of entomopathogenic fungi from Korean soils. Biocontrol Sci Technol. 23(3):288–304. doi: 10.1080/09583157.2012.756853.
- Sullivan CF, Parker BL, Skinner M. 2022. A review of commercial Metarhizium-and Beauveria-based biopesticides for the biological control of ticks in the USA. Insects. 13(3):260. doi: 10.3390/insects13030260.
- Sun HH, Zhang YJ, Zhang S. 2021. Complete mitogenome of the entomopathogenic fungus Metarhizium album and phylogenetic analysis of Hypocreales. Mitochondrial DNA Part B Resour. 6(6):1689–1690. doi: 10.1080/23802359.2021.1914229.
- Tillich M, Lehwark P, Pellizzer T, Ulbricht-Jones ES, Fischer A, Bock R, Greiner S. 2017. GeSeq–versatile and accurate annotation of organelle genomes. Nucleic Acids Res. 45(W1):W6–W11. doi: 10.1093/nar/gkx391.
- Wang L, Zhang S, Li JH, Zhang YJ. 2018. Mitochondrial genome, comparative analysis and evolutionary insights into the entomopathogenic fungus Hirsutella thompsonii. Environ Microbiol. 20(9):3393–3405. doi: 10.1111/1462-2920.14379.
- Zhang S, Ren LY, Zhang YJ. 2020a. Complete mitogenome of the entomopathogenic fungus Metarhizium rileyi. Mitochondrial DNA Part B Resour. 5(2):1494–1495. doi: 10.1080/23802359.2020.1742596.
- Zhang YJ, Yang XB, Zhang S. 2020b. Complete mitogenome of the entomopathogenic fungus Akanthomyces lecanii. Mitochondrial DNA Part B Resour. 5(1):1021–1022. doi: 10.1080/23802359.2020.1721349.
- Zimmermann G, Papierok B, Glare T. 1995. Elias Metschnikoff, Elie Metchnikoff or Ilya Ilich Mechnikov (1845-1916): a pioneer in insect pathology, the first describer of the entomopathogenic fungus Metarhizium anisopliae and how to translate a Russian name. Biocontrol Science and Technology. 5(4):527–530. doi: 10.1080/09583159550039701.
