Abstract
The mitogenome sequence of Hyperhalosydna striata was determined for the first time in the present study. The genome is 15,226 bp long and contains 13 protein-coding genes (PCGs), two ribosomal RNA genes (rRNAs), and 22 transfer RNA genes (tRNA). The overall base composition was 28.0% A, 21.9% C, 13.0% G, and 37.1% T. A phylogenetic tree was constructed to infer the phylogenetic position of H. striata among polynoid species whose mitochondrial genome sequences are available in GenBank. Hyperhalosydna striata was closely related to the species of subfamily Lepidonotinae.
Keywords:
Introduction
The scale worm, Hyperhalosydna striata (Kinberg Citation1856), belongs to the family Polynoidae Kinberg, Citation1856. It is easily distinguished from its congeners by the presence of stripes on the elytra. They are known to be free-living or to live in association with other polychaetes, such as eunicids (Hanley and Bruke Citation1991; Park et al. Citation2016). The type locality of H. striata is Jackson Port, Sydney, Australia (Kinberg Citation1856). However, this species is widely distributed throughout the South Pacific and the Indo-West Pacific, including Asian Waters (Grube Citation1876; McIntosh Citation1885; Moore Citation1903; Augener Citation1922; Fauvel Citation1932; Knox Citation1951; Imajima and Hartman Citation1964; Pillai Citation1965; Uschakov Citation1982; Uchida Citation1988; Imajima Citation2001; Wehe Citation2006; Park et al. Citation2016).
Recently, mitochondrial genomes have been used for phylogenetic and evolutionary studies of the highly diverse Polynoidae (Zhang et al. Citation2018; Gonzalez et al. Citation2021). However, studies are still scarce. For this reason, an additional complete mitogenome of the polynoid species H. striata was analyzed in this study.
Materials and methods
The specimen was collected by scuba diving in a subtidal rocky zone (depth 18 m) of Jejudo Island (33°13′41.29″N, 126°33′41.29″E, Munseom Islet, Seogwipo-si, South Korea) (). Species identification was performed under a stereomicroscope based on the description of Park et al. (Citation2016). The specimen was deposited at the National Institute of Biological Resources (NIBR, http://www.nibr.go.kr/, Eun-Jung Nam, [email protected]), Republic of Korea (NIBRIV0000902986). Genomic DNA was extracted from the pygidium of the specimen using the DNeasy Blood and Tissue kit (Qiagen, Hilden, Germany). REPLI-g Mitochondrial DNA Kit (Qiagen, Hilden, Germany) was used for mitochondrial DNA amplification, and mitochondrial genome sequencing was performed using the NovaSeq 6000 sequencing system (Illumina, San Diego, CA). Phylogenetic analysis was conducted to examine the phylogenetic position of H. striata using MEGA X software (Kumar et al. Citation2018). The tree was reconstructed using the ML method using the GTR + G + I model with 1000 bootstrap replicates. Illumina sequencing data of Hyperhalosydna striata were mapped on H. striata mitochondrial genome sequence and depth of mapped reads was calculated respectively using clc_ref_assemble and clc_mapping_info with default parameters in CLC Assembly Cell package ver. 4.2.1 (Qiagen, Aarhus, Denmark). Assembler and annotation tools, NOVOPlasty (Dierckxsens et al. Citation2017) and Chlorobox (Tillich et al. Citation2017), were used, respectively.
Results and discussion
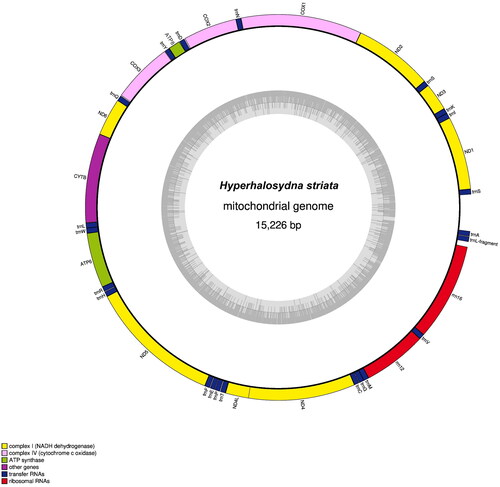
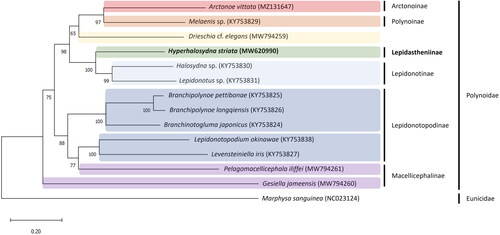
The complete mitogenome of H. striata (GenBank accession no. MW620990) was 15,226 bp long and consisted of 13 protein-coding genes (PCGs), two ribosomal RNA (rRNA) genes, and 22 transfer RNA (tRNA) genes (). The overall nucleotide composition was 28.0% A, 21.9% C, 13.0% G, and 37.1% T, with high G + C content (65.1%). The average mapping depth is ×410,057 (Figure S1). The phylogenetic tree results showed that H. striata sister was cluster to the Lepidonotinae species (Halosydna sp., Lepidonotus sp.) ().
Figure 3. Maximum-likelihood (ML) tree reconstructed using a concatenated data set of 13 protein-coding genes based on 14 mitogenome sequences, including Hyperhalosydna striata from the present study. Bootstrap replicates were performed 1000 times. The GenBank accession number of each species is shown in parentheses after the species name. The following sequences were used: Hyperhalosydna striata MW620990, Arctonoe vittata MZ131647 (Park et al. Citation2021), Melaenis sp. KY753829 (Zhang et al. Citation2018), Drieschia cf. elegans MW794259 (Gonzalez et al. Citation2021), Halosydna sp. KY753830 (Zhang et al. Citation2018), Lepidonotus sp. KY753831 (Zhang et al. Citation2018), Branchipolynoe pettibonae KY753825 (Zhang et al. Citation2018), B. longqiensis KY753826 (Zhang et al. Citation2018), Branchinotogluma japonicus KY753824 (Zhang et al. Citation2018), Lepidonotopodium okinawae KY753838 (Zhang et al. Citation2018), Levensteiniella iris KY753827 (Zhang et al. Citation2018), Pelagomacellicephala iliffei MW794261 (Gonzalez et al. Citation2021), and Gesiella jameensis MW794260 (Gonzalez et al. Citation2021).
Ethical approval
The material of this paper does not involve ethical conflicts. Hyperhalosydna striata is neither endangered on the CITES catalogue nor collected from a natural reserve.
Author contributions
Kwang-Soo Kim conducted collecting the specimen, species identification, and wrote draft. Jiseon Park analyzed the mitogenome sequence and wrote draft. Taeseo Park contributed conception, designing this study, and revising manuscript.
Supplemental Material
Download PNG Image (45.6 KB)Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The genome sequence data supporting this study’s findings are available in the National Center for Biotechnology Information (NCBI) GenBank (https://www.ncbi.nlm.nih.gov) under accession no. MW620990. The associated BioProject, SRA, and BioSample numbers were PRJNA727906, SRR14566003, and SAMN19229862, respectively. The data that support the findings of this study are openly available in Mendeley (https://doi.org/10.17632/9h4gtkx9fn.1).
Additional information
Funding
References
- Augener H. 1922. Revision der Australischen Polychaeten-typen von Kinberg. Arkiv Zool. 14(8):1–42.
- Dierckxsens N, Mardulyn P, Smits G. 2017. NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 45(4):e18. doi:10.1093/nar/gkw955.
- Fauvel P. 1932. Annelida Polychaeta of the Indian Museum, Calcutta. Mem Indian Mus. 12(1):1–262.
- Gonzalez BC, Martínez A, Worsaae K, Osborn KJ. 2021. Morphological convergence and adaptation in cave and pelagic scale worms (Polynoidae, Annelida). Sci Rep. 11(1):10718. doi:10.1038/s41598-021-89459-y.
- Grube AE. 1876. Bemerkungen über die Familie der aphroditeen (Gruppe Polynoina, Acoëta, Polylepidea). Jahresber Schlesische Gesellschaft Vaterländische Cultur Breslau. 53:46–72.
- Hanley JR, Bruke M. 1991. Polychaeta Polynoidae: scaleworms of the Chesterfield Island and Fairway Reefs, Coral Sea. Mém Mus Natl d’Hist Nat Paris Sér A. 151:9–82.
- Imajima M. 2001. Annelida, Polychaeta II. Tokyo, Japan: Seibutsu Kenkyujo Publishing Co.; p. 1–542.
- Imajima M, Hartman O. 1964. The polychaetous annelids of Japan. Los Angeles (CA): University of California Press; p. 1–452.
- Kinberg JGH. 1856. Nya slägten och arter af Annelider, Öfversigt af Kongl. Vetenskaps-Akad Förhhandl Stockh. 12(9–10):381–388.
- Knox GA. 1951. The polychaetous annelids of Banks Peninsula. Rec Canterb Mus. 5(5):312–329.
- Kumar S, Stecher G, Li M, Knyaz C, Tamura K. 2018. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. 35(6):1547–1549. doi:10.1093/molbev/msy096.
- McIntosh WC. 1885. Report on the Annelida Polychaeta collected by H.M.S. challenger during the years 1873–1876. Report on the scientific results of the voyage of H.M.S. challenger during the years 1873–76. Zoology. Vol. 12; p. 1–554.
- Moore JP. 1903. Polychaeta from the coastal slope of Japan and from Kamchatka and Bering Sea. Proc Acad Nat Sci Philadelph. 55:401–490.
- Park J, Jung J, Kim K-S, Park T. 2021. Complete mitochondrial genome of the commensal scale worm, Arctonoe vittata (Grube, 1855) (Polychaeta: Polynoidae), collected from benthic habitat of the eastern coast of Korea. Mitochondrial DNA Part B. 6(8):2455–2457. doi:10.1080/23802359.2021.1955771.
- Park T, Lee SK, Kim W. 2016. New record of scale worms, Arctonoe vittata (Grube, 1855) and Hyperhalosydna striata (Kinberg, 1856) (Polychaeta: Polynoidae) from Korean waters. J Spec Res. 5(3):517–529. doi:10.12651/JSR.2016.5.3.517.
- Pillai TG. 1965. Annelida Polychaeta from the Philippines and Indonesia. Ceyl J Sci (Biol Sci). 5(2):110–177.
- Tillich M, Lehwark P, Pellizzer T, Ulbricht-Jones ES, Fischer A, Bock R, Greiner S. 2017. GeSeq – versatile and accurate annotation of organelle genomes. Nucleic Acids Res. 45(W1):W6–W11. doi:10.1093/nar/gkx391.
- Uchida H. 1988. Polychaeta fauna in Wakayama Prefecture. Nanki Seibutsu. 30(2):75–86.
- Uschakov PV. 1982. Polychaetes of the suborder Aphroditiformia of the Arctic Ocean and the Northwestern Part of the pacific, families Aphroditidae and Polynoidae. FAUNA SSSR, Mnogoshchetinkovyye Chervil (fauna of the USSR, Polychaeta). Moscow: Academiya Nauk SSSR; p. 1–272.
- Wehe T. 2006. Revision of the scale worms (Polychaeta: Aphroditoidea) occurring in the seas surrounding the Arabian Peninsula. Part I: Polynoidae. Fauna Arabia. 22:23–197.
- Zhang Y, Sun J, Rouse GW, Wiklund H, Pleijel F, Watanabe HK, Chen C, Qian PY, Qiu JW. 2018. Phylogeny, evolution and mitochondrial gene order rearrangement in scale worms (Aphroditiformia, Annelida). Mol Phylogenet Evol. 125:220–231. doi:10.1016/j.ympev.2018.04.002.