Abstract
Eremurus zoae Vved. 1971 is a perennial herbaceous plant in the family Asphodelaceae and an endemic species of the Kyrgyz Republic; however, its complete chloroplast genome sequence has not been reported. Here, we investigated the complete chloroplast (cp) genome of E. zoae using next-generation sequencing. The cp genome was 153,744 bp long, with a large single copy (84,020 bp), a small single copy (16,766 bp), and a pair of inverted repeats (26,479 bp). The genome encodes 132 genes, including 86 protein-coding genes, 38 tRNA genes, and 8 rRNA genes. Phylogenetic analysis revealed that the genus Eremurus forms a monophyletic group and E. zoae is closely related to E. chinensis. This study provides a molecular foundation for future phylogenetic studies of Eremurus.
Introduction
Eremurus M.Bieb. (Asphodelaceae) is known as foxtail lilies or desert candles, which is a perennial herbaceous plant belonging to the Asphodelaceae family (APG IV Citation2016; Muhidinov et al. Citation2020; Farhadi et al. Citation2023). This genus, which comprises over 50 species, is mainly distributed in Central and Western Asia, with the center of its diversity in Central Asia (Hedge and Wendelbo Citation1963; Naderi Safar et al. Citation2009; Farhadi et al. Citation2023). It is characterized by swollen, fleshy, thick roots; leafless flowering stems; white to pink or yellow flowers; and campanulate or funnel-shaped perianths (Fedchenko Citation1968; Xinqi et al. Citation2000; Naderi Safar et al. Citation2009; Farhadi et al. Citation2023). Modern pharmacological research has shown that many species in this genus exhibit antibacterial, antimicrobial, antioxidant, anti-inflammatory, antiradical, and anti-glycation activities (Karaman et al. Citation2011; Zhu et al. Citation2013). Eremurus is also an economically important plant with edible, medicinal, and ornamental uses (Kamenetsky and Rabinowitch Citation1999; Naderi Safar et al. Citation2009; Safar et al. Citation2014; Farhadi et al. Citation2023). Additionally, several Eremurus species contain water-soluble glucomannans and galactomannans in their roots. These substances become sticky when in contact with water and are utilized in the form of glue (Makhmudjanov et al. Citation2019; Muhidinov et al. Citation2020). Eremurus zoae Vved. 1971, which is endemic to Kyrgyz Republic, can be distinguished from related species by its 25–40 cm tall (), rather short and dense conical-cylindrical raceme with yellow flowers (). It is also often misidentified as Eremurus luteus Baker, but this species is clearly different from E. luteus by the shape of its capsule (): E. luteus has an elongated capsule, whereas E. zoae has a globose capsule (Lazkov and Sennikov Citation2015). This species also faces threats from factors such as overgrazing and plant collection. Consequently, it was classified as vulnerable (VU) in the Red Data Book of the Kyrgyz Republic (Citation2006). Therefore, investigating the genetic diversity and structure of this species to conserve its genetic resources is essential. Although the chloroplast (cp) genome is a very useful genetic tool for inferring phylogenetic relationships, few studies involving the complete cp genome of Eremurus have been published (Cho et al. Citation2015; Song et al. Citation2022). Here, we report the complete cp genome of E. zoae for the first time to clarify the phylogenetic relationships between E. zoae and related species and to facilitate further research for a systematic understanding of Eremurus.
Figure 1. Eremurus zoae Vved 1971 (Photographs was taken by Georgy A. Lazkov in Issyk Kul, Kyrgyz Republic). A, Habitat; B, Flower; C, Capsule. E. zoae is a perennial herb with a height of 25–40 cm. The flowers are campanulate, with yellow petals that reflex in fruiting. The capsules are globose. The flowering period is from April to May and the fruiting period is from May to June.

Materials and methods
Eremurus zoae was collected from Issyk Kul, Kyrgyz Rupublic (latitude 42°27′32.4″, longitude 76°05′42.0″; ). The voucher specimen was deposited at the herbarium of the Korea National Arboretum (KH, http://www.nature.go.kr/kbi/plant/smpl/KBI_2001_030100.do, Hee-Young Gil, E-mail: [email protected]) under the voucher number KZ_160513_403. Fresh leaves were dried using silica gel. DNA was extracted from the silica gel-dried leaves using a DNeasy Plant Mini Kit (Qiagen Inc., Valencia, CA, USA). Next-generation sequencing was conducted on the MiSeq platform (Illumina Inc., Seoul, South Korea) with a 500 bp insert size, and 9,009,222 reads were obtained. We performed de novo assembly using GetOrganelle (Jin et al. Citation2020) with E. chinensis as a reference (NC_070056) and conducted the final assembly of the complete cp genome using the Geneious Prime program (Kearse et al. Citation2012). The cp genome was annotated using GeSeq (Tillich et al. Citation2017) and the tRNAscan-SE software (Lowe and Chan Citation2016). A circular map of the E. zoae cp genome and cis- and trans-spliced genes was generated using CPGview (Liu et al. Citation2023). The complete cp genome of E. zoae was submitted to GenBank under the accession number OR264467.
To investigate the phylogenetic relationships, we analyzed 17 species together with three outgroup taxa (one Xeronemataceae and two Iridaceae), which were downloaded from NCBI GenBank, except E. zoae. For phylogenetic analysis, 82 protein-coding genes were aligned using MAFFT, and the most suitable model for the maximum likelihood (ML) and Bayesian inference (BI) methods was determined using ModelFinder in the PhyloSuite software (Katoh and Standley Citation2013; Kalyaanamoorthy et al. Citation2017; Zhang et al. Citation2020). The ML tree was constructed using IQ-TREE under the GTR + R3 + F model with 5,000 ultrafast bootstraps, whereas the BI tree was inferred using MrBayes 3.2.6 under the GTR + F + I + G4 model (two parallel runs, 2,000,000 generations), in which the initial 25% of the sampled data were discarded as burn-in (Guindon et al. Citation2010; Ronquist et al. Citation2012; Minh et al. Citation2013; Nguyen et al. Citation2015; Zhang et al. Citation2020).
Results and discussion
The cp genome of E. zoae was 153,744 bp in length, with an overall GC content of 42.9% and an average coverage of 446.60 (, Figure S1). The assembled genome has a typical quadripartite structure, including a large single copy (LSC: 84,020 bp), a small single copy (SSC: 16,766 bp), and a pair of inverted repeats (IRa and IRb: 26,479 bp). The cp genome contained 132 genes (86 protein-coding genes, 38 tRNA genes, and 8 rRNA genes), including 20 genes (8 protein-coding genes, 8 tRNA genes, and 4 rRNA genes) duplicated in the IR regions. In total, 11 cis-splicing genes (Figure S2) and one trans-splicing gene, rps12 (Figure S3), were detected. Of the 11 cis-splicing genes, nine (rps16, atpF, rpoC1, petB, petD, rpl16, rpl2, ndhB, and ndhA) contained one intron, and two (pafl, clpP1) contained two introns. Gene rpl32 was missing in some species of subfamily Asphodelaceae, which is related to Eremurus zoae (including genus Eremurus, Aloe, Aloidendron in ). The absence of rpl32 gene has been recently reported in Aloe vera (Xie et al. Citation2019). Gene losses in plastid genomes might be attributed to the functional transfer to the nucleus or replacement of nuclear genes (Shrestha et al. Citation2020; Han et al. Citation2022).
Figure 2. Complete chloroplast genome map of Eremurus zoae Vved. drawn using CPGview. The circle map contains six tracks. The first track indicates the repeat distribution. The second track shows the tandem repeats as a blue bar. The third track represents microsatellite sequences in green and yellow. The fourth track shows the LSC, SSC, and IR regions. The fifth track indicates the GC contents, and the sixth track represents genes.
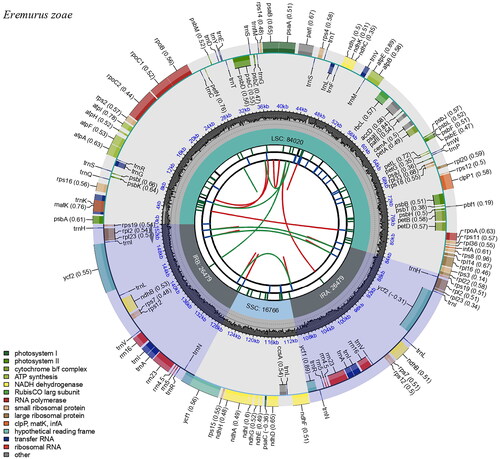
Figure 3. Phylogenetic tree of 17 species with three outgroup taxa (one Xeronemataceae, two Iridaceae) based on 82 protein-coding genes using the ML and BI methods. The numbers above the nodes indicate the bootstrap support values and the Bayesian posterior probabilities. The newly reported chloroplast genome in this study is indicated in red. All sequences used in the analysis are as follows: Eremurus zoae (OR264467, this study), Eremurus chinensis (NC070056), Eremurus robustus (NC046772, Makhmudjanov et al.Citation2019), Aloe vera (NC035506), Aloe maculata (NC035505), Aloidendron pillansii (NC044761), Dianella nigra (MN239902), Xanthorrhoea preissii (NC035996), Theropogon pallidus (NC070131), Asparagus officinalis (NC034777), Scilla siberica (NC061320), Milla biflora (NC036000), Aphyllanthes monspeliensis (NC035968), Amaryllis belladonna (MZ433380), Xeronema callistemon (NC072587, Kamra et al.Citation2023), Moraea spathulata (NC072577, Kamra et al.Citation2023), and Iris petrana (NC063920, Volis et al.Citation2022).
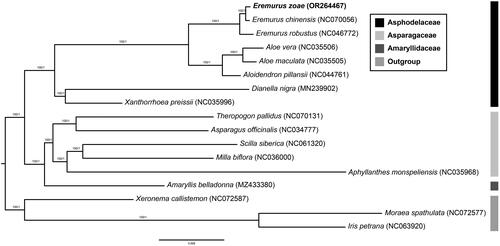
The topologies of the two methods (ML and BI) precisely correspond. Phylogenetic trees strongly supported the idea that Asphodelaceae is a monophyletic group (bootstrap value = 100). Within Asphodelaceae, three subfamilies (Asphodeloideae, Xanthorrhoea, and Hemerocallidoideae) formed distinct clades with high support values. E. zoae was the most closely related to E. chinensis (). Our results are consistent with those of a previous Asparagales phylogenetic study (Chase et al. Citation2009).
Conclusions
In this study, we reported the first complete chloroplast genome of E. zoae. The plastid genome of E. zoae was determined to be 153,744 bp in length, containing a total of 132 genes. A phylogenetic tree reconstructed by concatenated 82 protein-coding genes showed that Eremurus formed a monophyletic group. Our tree also revealed that E. zoae was most closely related to E. chinensis. This result provides useful information regarding the phylogenetic system and evolution of the genus Eremurus.
Ethical approval
As the collected plant samples were not endangered or protected, no specific permits were required. This study was conducted in compliance with the relevant institutional, national, and international guidelines and legislation.
Authors’ contribution
HY Gil designed the study and revised the manuscript. HJ Jeong, AL Kim, and YR Choi performed the experiments. GA Lazkov, CG Jang, and HJ Choi collected plant materials. JE Jang analyzed the data and drafted the manuscript. All the authors have read and agreed to the submitted version of the manuscript.
Supplemental Material
Download MS Word (481.7 KB)Disclosure statement
No potential conflict of interest was reported by the authors.
Data availability statement
The genome sequence data supporting the findings of this study are openly available in GenBank of NCBI at [https://www.ncbi.nlm.nih.gov] (https://www. ncbi. nlm. nih. gov) under accession no. OR264467. The associated BioProject, SRA, and Bio-Sample numbers are PRJNA996712, SRR25367042, and SAMN36621056, respectively.
Additional information
Funding
References
- APG IV. 2016. An update of the Angiosperm Phylogeny Group classification for the orders and families of flowering plants: APG IV. Bot J Linn Soc. 181(1):1–20. doi:10.1111/boj.12385.
- Chase MW, Reveal JL, Fay MF. 2009. A subfamilial classification for the expanded asparagalean families Amaryllidaceae, Asparagaceae and Xanthorrhoeaceae. Bot J Linn Soc. 161(2):132–136. doi:10.1111/j.1095-8339.2009.00999.x.
- Cho KS, Yun BK, Yoon YH, Hong SY, Mekapogu M, Kim KH, Yang TJ. 2015. Complete chloroplast genome sequence of tartary buckwheat (Fagopyrum tataricum) and comparative analysis with common buckwheat (F. esculentum). PLoS One. 10(5):e0125332. doi:10.1371/journal.pone.0125332.
- Farhadi F, Avan R, Sahebkar A, Eghbali S. 2023. A comprehensive review on Eremurus species: phytochemistry, pharmacology and traditional uses. Phytochem Lett. 53:142–149. doi:10.1016/j.phytol.2022.12.002.
- Fedchenko BA. 1968. Eremurus. In: komarov VL, editor. Flora of the USSR. Vol. 4. Leningrad: botanical Institute of Academy of Science; p. 27–40.
- Guindon S, Dufayard JF, Lefort V, Anisimova M, Hordijk W, Gascuel O. 2010. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol. 59(3):307–321. doi:10.1093/sysbio/syq010.
- Han S, Ding H, Bi D, Zhang S, Yi R, Gao J, Yang J, Ye Y, Wu L, Kan X. 2022. Structural Diversities and Phylogenetic Signals in Plastomes of the Early-Divergent Angiosperms: A Case Study in Saxifragales. Plants. 11(24):3544. doi:10.3390/plants11243544.
- Hedge I, Wendelbo P. 1963. Notes on the giant Asphodels of Afghanistan. J Roy Hort Soc London. 88(9):402–406.
- Jin JJ, Yu WB, Yang JB, Song Y, dePamphilis CW, Yi TS, Li DZ. 2020. GetOrganelle: a fast and versatile toolkit for accurate de novo assembly of organelle genomes. Genome Biol. 21(1):241. doi:10.1186/s13059-020-02154-5.
- Kalyaanamoorthy S, Minh BQ, Wong TK, Von Haeseler A, Jermiin LS. 2017. ModelFinder: fast model selection for accurate phylogenetic estimates. Nat Methods. 14(6):587–589. doi:10.1038/nmeth.4285.
- Kamenetsky R, Rabinowitch E. 1999. Flowering response of Eremurus to post-harvest temperatures. Sci Hortic. 79(1-2):75–86. doi:10.1016/S0304-4238(98)00181-2.
- Kamra K, Jung J, Kim JH. 2023. A phylogenomic study of Iridaceae Juss. based on complete plastid genome sequences. Front Plant Sci. 14:1066708. doi:10.3389/fpls.2023.1066708.
- Karaman K, Polat B, Ozturk I, Sagdic O, Ozdemir C. 2011. Volatile compounds and bioactivity of Eremurus spectabilis (Ciris), a Turkish wild edible vegetable. J Med Food. 14(10):1238–1243. doi:10.1089/jmf.2010.0262.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780. doi:10.1093/molbev/mst010.
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, et al. 2012. Geneious basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 28(12):1647–1649. doi:10.1093/bioinformatics/bts199.
- Kyrgyz Republic. 2006. The Red Data Book of Kyrgyz Republic. 2nd ed. Bishkek: kyrgyz Republic; p. 543.
- Lazkov G, Sennikov AN. 2015. Taxonomic corrections and new records in vascular plants of Kyrgyzstan, 4. Memoranda Societatis Pro Fauna et Flora Fennica. 91:67–83.
- Liu S, Ni Y, Li J, Zhang X, Yang H, Chen H, Liu C. 2023. CPGView: a package for visualizing detailed chloroplast genome structures. Mol Ecol Resour. 23(3):694–704. doi:10.1111/1755-0998.13729.
- Lowe TM, Chan PP. 2016. TRNAscan-SE online: integrating search and context for analysis of transfer RNA genes. Nucleic Acids Res. 44(W1):W54–W57. doi:10.1093/nar/gkw413.
- Makhmudjanov D, Yusupov Z, Abdullaev D, Deng T, Tojibaev K, Sun H. 2019. The complete chloroplast genome of Eremurus robustus (Asphodelaceae). Mitochondrial DNA B Resour. 4(2):3366–3367. doi:10.1080/23802359.2019.1674198.
- Minh BQ, Nguyen MA, von Haeseler A. 2013. Ultrafast approximation for phylogenetic bootstrap. Mol Biol Evol. 30(5):1188–1195. doi:10.1093/molbev/mst024.
- Muhidinov ZK, Bobokalonov JT, Ismoilov IB, Strahan GD, Chau HK, Hotchkiss AT, Liu L. 2020. Characterization of two types of polysaccharides from Eremurus hissaricus roots growing in Tajikistan. Food Hydrocolloids. 105:105768. doi:10.1016/j.foodhyd.2020.105768.
- Naderi Safar K, Kazempour Osaloo S, Zarrei M. 2009. Phylogeny of the genus Eremurus (Asphodelaceae) based on morphological characters in the Flora Iranica area. Iran J Bot. 15(1):27–35.
- Nguyen LT, Schmidt HA, von Haeseler A, Minh BQ. 2015. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 32(1):268–274. doi:10.1093/molbev/msu300.
- Ronquist F, Teslenko M, van der Mark P, Ayres DL, Darling A, Höhna S, Larget B, Liu L, Suchard MA, Huelsenbeck JP. 2012. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol. 61(3):539–542. doi:10.1093/sysbio/sys029.
- Safar KN, Osaloo SK, Assadi M, Zarrei M, Mozaffar MK. 2014. Phylogenetic analysis of Eremurus, Asphodelus, and Asphodeline (Xanthorrhoeaceae-Asphodeloideae) inferred from plastid trnL-F and nrDNA ITS sequences. Biochem Syst Ecol. 56:32–39. doi:10.1016/j.bse.2014.04.015.
- Shrestha B, Gilbert LE, Ruhlman TA, Jansen RK. 2020. Rampant nuclear transfer and substitutions of plastid genes in Passiflora. Genome Biol Evol. 12(8):1313–1329. doi:10.1093/gbe/evaa123.
- Song W, Ji C, Chen Z, Cai H, Wu X, Shi C, Wang S. 2022. Comparative analysis the complete chloroplast genomes of nine Musa Species: genomic features, comparative analysis, and phylogenetic implications. Front Plant Sci. 13:832884. doi:10.3389/fpls.2022.832884.
- Tillich M, Lehwark P, Pellizzer T, Ulbricht-Jones ES, Fischer A, Bock R, Greiner S. 2017. GeSeq–versatile and accurate annotation of organelle genomes. Nucleic Acids Res. 45(W1):W6–W11. doi:10.1093/nar/gkx391.
- Volis S, Zhang Y, Deng T, Yusupov Z. 2022. Dark-colored Oncocyclus irises in Israel analyzed by AFLP, whole chloroplast genome sequencing and species distribution modeling. Israel J Ecol Evol. 68(1–4):43–53. doi:10.1163/22244662-bja10037.
- Xie DF, Yu HX, Price M, Xie C, Deng YQ, Chen JP, Yu Y, Zhou SD, He XJ. 2019. Phylogeny of Chinese Allium species in section Daghestanica and adaptive evolution of Allium (Amaryllidaceae, Allioideae) species revealed by the chloroplast complete genome. Front Plant Sci. 10:460. doi:10.3389/fpls.2019.00460.
- Xinqi C, Songyun L, Jiemei X, Tamura MN. 2000. Liliaceae. Flora of China. In: Wu ZY, Raven PH, editors. Flora of China. Vol 24. Beijing and St. Louis (MO): Science Press and Missouri Botanical Garden Press. p. 73–263.
- Zhang D, Gao F, Jakovlić I, Zou H, Zhang J, Li WX, Wang GT. 2020. PhyloSuite: an integrated and scalable desktop platform for streamlined molecular sequence data management and evolutionary phylogenetics studies. Mol Ecol Resour. 20(1):348–355. doi:10.1111/1755-0998.13096.
- Zhu Y, Xu C-h, Huang J, Li G-y, Zhou Q, Liu X-H, Sun S-q, Wang J-h 2013. Rapid discrimination of three Uighur medicine of Eremurus by FT-IR combined with 2DCOS-IR. J Mol Struct. 1069:96–102. doi:10.1016/j.molstruc.2013.11.054.
