Abstract
In the present study, the complete mitochondrial genome (mitogenome) of the Papilio macilentus (Lepidoptera: Papilionoidea: Papilionidae) was sequenced by next-generation sequencing method. The mitochondrial genome is a circular DNA molecule of 15,264 bp in size with 80.7% AT content, including 37 genes (13 protein-coding genes, 2 rRNA genes, and 22 tRNA genes), and a long non-coding region (Control region). All protein-coding genes are initiated by ATN codons, and terminated with TAA, TAG, or single T. All tRNAs can be folded into common clover leaf secondary structure, except trn-S1. Phylogenetic analyses based on 13 protein-coding genes and 2 rRNA genes using maximum likelihood and Bayesian inference confirmed that P. macilentus and Papilio memnon are clustered into a clade, and revealed the relationships between Papilionini, Troidini, Teinopaippini and Leptocircini.
Introduction
Papilionidae has attracted more attention due to extensive morphological diversity and their colorful wing patterns, and have been widely studied regarding ecological adaption, phylogeny, genetics, and evolution (He et al. Citation2022). Papilio macilentus Janson, 1877 belongs to Papilionidae, and is mainly distributed in China, Korea, Japan, and Eastern Siberia, more than one or two generations a year in China, larvae mainly feed on Zanthoxylum and Ruta, and adults are visible from April to August in Anhui province, China (Wu Citation2001). As a member of the ornamental butterfly species, P. macilentus is morphologically similar to Papilio bianor, but P. macilentus shows more narrow hind wings, and the outer margin area is different in red spots (). Relatively little attention has been paid to P. macilentus, possibly because the population of the P. macilentus is significantly smaller in nature. In order to fill in the gap and clarify the mitochondrial genome (mitogenome) characteristics of P. macilentus, we have sequenced and annotated the complete mitogenome of P. macilentus by next-generation sequencing method, which would be useful for species identification, phylogenetic analysis and conservation of this species.
Materials and methods
In this study, total genome DNA was extracted from the male adult leg of P. macilentus which was collected from Chuzhou City (118.295 E, 32.271 N), Anhui Province, China in April 2017. The total DNA was extracted using a DNeasy© Tissue Kit (Qiagen, Hilden, Germany). The genome was sequenced by the next-generation sequencing method with the Illumina Hiseq 2500 platform. The mitogenome was assembled using Geneious Primer v.2021.2.2 (Kearse et al. Citation2012) and annotated with MITOS server (Bernt et al. Citation2013) and NCBI BLAST (https://blast.ncbi.nlm.nih.gov). The specimen and genome DNA were deposited at −80 °C in the Animal Collection of Chuzhou University with the voucher number CHZU-LPP-0254 (Yan Dong, [email protected]). To investigate the phylogenetic status of P. macilentus in Papiliononae, 46 currently available complete mitogenomes of Papiliononae retrieved from GenBank were used in phylogenetic analysis, two Parnassiinae insects (Sericinus montela, HQ259122 and Zerynthia polyxena, MK507888) were used as outgroup. The phylogenetic tree was reconstructed with maximum likelihood (ML) and Bayesian inference (BI) methods based on the 13 protein-coding genes (PCGs) and 2 rRNAs by IQ-TREE v2.1.3 (Minh et al. Citation2020) and PhyloBayes MPI 1.8c (Lartillot et al. Citation2013), respectively. The number of bootstrap replicates was set to 1000 with automatic model prediction in ML analyses. The site-heterogeneous mixture model (CAT + GTR) was implemented in BI analyses. The consensus tree was computed with a burn-in of 25% of sampled values. Each gene was aligned using MAFFT 7 (Katoh K & Standley, Citation2013) and trimmed with trimAl 1.4.1 (Capella-Gutiérrez et al. Citation2009), then concatenated individual genes using MEGA X (Kumar et al. Citation2018).
Results
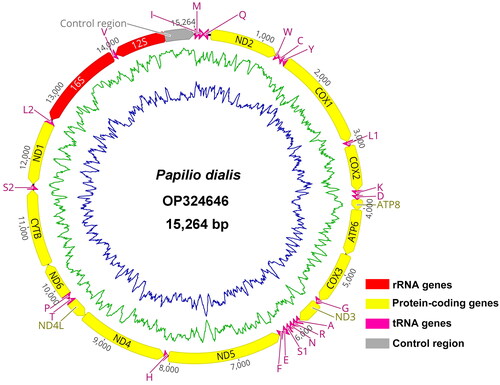
In this research, about 6.4 Gb of clean data were obtained and produced a final mitogenome for P. macilentus with an average sequencing depth of 1,374× (Figure S1). After checking and annotating, the complete mitogenome of P. macilentus is 15,264 bp (), which is comparable to the sizes of previously documented mitogenomes of Papiliononae species, ranging from 14,642 bp of Graphium antiphates (MN013005) to 16,094 bp of Papilio maraho (FJ810212; Wu et al. Citation2010; Liu et al. Citation2020), the gene order of the P. macilentus mitogenome is identical to other commonly sequenced Papilionidae species. The mitogenome of P. macilentus nucleotide composition was A (39.7%), T (41.0%), G (7.4%), C (11.9%), and A + T content (80.7%). The complete mitogenome was composed of 37 typical mitochondrial genes (13 PCGs, 22 transfer RNA (tRNA) genes, two ribosomal RNA (rRNA) genes), and one large non-coding region (Control region). The total length of the 13PCGs is 11,216 bp, and these encode 3,727 amino acids, accounting for 73.5% of the complete mitogenome. All the PCGs are initially encoded by ATN and terminated coding with TAA, TAG, or single T. The base composition of the 13 PCGs is 79.4 A + T (A = 33.6%, T = 45.8%, C = 10.1%, and G = 10.5%).
All of the tRNAs were identified by ARWEN version 1.2 software (Laslett and Canbäck Citation2008), all tRNAs could form a typical cloverleaf structure except trn-S1, in which the dihydrouridine arm formed a loop. The lengths of the tRNAs ranged from 60 bp (trrn-S1) to 71 bp (trn-K). rRNA genes are identified by comparison with Papilio memnon (MN013052). The 16S rRNA gene is 1,322 bp in size and is located between trn-L1 and trn-V; the 12S rRNA gene is 772 bp in length and is located after trn-V. The control region is located between 12S rRNA and trn-I with 457 bp.
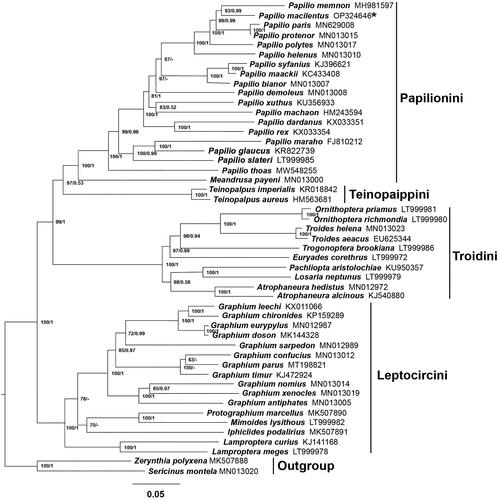
The resulting tree was represented and edited using FigTree v1.4.1 (Figures S2 and S3). The two topologies are highly similar (). Both trees support the monophyly of each tribe of Papiliononae with high bootstrap among almost all nodes. Papilionini and Teinopaippini were sister groups and then clustered with Troidini and eventually converged with Leptocircini. The result also indicated that P. macilentus has the closest relationship with P. memnon and is clustered within Papilionini clade.
Figure 3. Phylogenetic tree of Papilio macilentus and other related species based on 13 PCGs and 2 rRNAs. The numbers at the nodes separated by ‘/’ indicate analyses based on either the ML (left) or BI (right) methods, respectively. ‘-’ are lack of Bayesian posterior probabilities indicates that the nodes were not recovered in the BI analysis. GenBank accession numbers of each mitogenome are given after the species name, and the asterisk indicates the species sequenced in this study. The following sequences were used: Zerynthia polyxena MK507888 (Condamine et al. Citation2018); Troides helena MN013023 (Liu et al. Citation2020); Troides aeacus EU625344; Trogonoptera brookiana LT999986; Teinopalpus imperialis KR018842 (Huang et al. Citation2015); Teinopalpus aureus HM563681 (Qin et al. Citation2012); Sericinus montela MN013020 (Liu et al. Citation2020); Protographium marcellus MK507890 (Condamine et al. Citation2018); Papilio xuthus KU356933 (Wu et al. Citation2016); Papilio thoas MW548255 (Jeng et al. Citation2021); Papilio syfanius KJ396621 (Dong et al. Citation2016); Papilio slateri LT999985; Papilio rex KX033354; Papilio protenor MN013015 (Liu et al. Citation2020); Papilio polytes MN013017 (Liu et al. Citation2020); Papilio paris MN629008 (Sun et al. Citation2020); Papilio memnon MH981597 (Shi et al. Citation2020); Papilio maraho FJ810212 (Wu et al. Citation2010); Papilio machaon HM243594; Papilio maackii KC433408 (Dong et al. Citation2013); Papilio helenus MN013010 (Liu et al. Citation2020); Papilio glaucus KR822739 (Cong et al. Citation2015); Papilio demoleus MN013008 (Liu et al. Citation2020); Papilio dardanus KX033351; Papilio bianor MN013007 (Liu et al. Citation2020); Pachliopta aristolochiae KU950357; Ornithoptera richmondia LT999980; Ornithoptera priamus LT999981; Mimoides lysithous LT999982; Meandrusa payeni MN013000 (Liu et al. Citation2020); Losaria neptunus LT999979; Lamproptera meges LT999978; Lamproptera curius KJ141168 (Qin et al. Citation2015); Iphiclides podalirius MK507891 (Condamine et al. Citation2018); Graphium xenocles MN013019 (Liu et al. Citation2020); Graphium timur KJ472924 (Chen et al. Citation2016a); Graphium sarpedon MN012989 (Liu et al. Citation2020); Graphium parus MT198821 (Duan et al. Citation2020); Graphium nomius MN013014 (Liu et al. Citation2020); Graphium leechi KX011066; Graphium eurypylus MN012987 (Liu et al. Citation2020); Graphium doson MK144328 (Kong et al. Citation2019); Graphium confucius MN013012 (Liu et al. Citation2020); Graphium chironides KP159289 (Chen and Hao Citation2016); Graphium antiphates MN013005 (Liu et al. Citation2020); Euryades corethrus LT999972; Atrophaneura hedistus MN012972 (Liu et al. Citation2020); Atrophaneura alcinous KJ540880 (Chen et al. Citation2016b).
Discussion and conclusion
In this study, the composition and structure of P. macilentus mitogenome are predicted and analyzed, and the features are consistent with those of other Papilio insects, including the general genome size, AT content, and gene arrangement (Zuo et al. Citation2018). ML and BI analysis produced two consensus structures on tribe level, with the topology (((Papilionini + Teinopaippini)+Troidini)+Leptocircini), and this topology was affirmed by Miller’s research (Citation1987) but in contrast to the later hypothesis, which believed that the Papilionini and Troidini formed a sister relationship, and then clustered with Teinopaippini (Simonsen et al. Citation2011). The monophyly of the tribe Papilionini was recovered in this research, the sister relationship of Papilio and Meandrusa has also been confirmed in previous studies (Yan et al. Citation2022). The phylogenetic status of Papilio demoleus in Papilionini and Iphiclides podalirius in Leptocircini is unclear. Otherwise, the relationships among Graphium parus, G. confucius, and G. timur were inconsistent by ML and BI analysis. These inconsistent analysis results may be due to insufficient sample size and the limited information sites of mitochondrial genes. This research will provide basic data for phylogenetic analysis and population genetic diversity protection of Papilionidae in the future.
Author contributions
YFW: conceptualization, analysis, interpretation of the data, and writing – original draft. WHY and AJ: formal analysis and editing. YD and JJW: investigation and formal analysis. LXZ: funding acquisition and writing – review. All authors agree to be accountable for all aspects of the work.
Ethical approval
Experiments were performed in accordance with the recommendations of the Ethics Committee of the College of Biology and Food Engineering, Chuzhou University. These policies were enacted according to the Chinese Association for the Laboratory Animal Sciences and the Institutional Animal Care and Use Committee (IACUC) protocols.
Supplemental Material
Download MS Word (2.1 MB)Disclosure statement
The authors have no conflicts of interest to report. The authors alone are responsible for the content and writing of the paper.
Data availability statement
The mitochondrion genome sequence data that support the findings of this study are openly available in GenBank of the National Center for Biotechnology Information (NCBI) at https://www.ncbi.nlm.nih.gov/ under the accession no. OP324646. The associated BioProject, Bio-Sample numbers, and SRA are PRJNA1044041, SAMN38366985, and SRR26915771, respectively.
Additional information
Funding
References
- Bernt M, Donath A, Jühling F, Externbrink F, Florentz C, Fritzsch G, Pütz J, Middendorf M, Stadler PF. 2013. MITOS: improved de novo metazoan mitochondrial genome annotation. Mol Phylogenet Evol. 69(2):313–319. doi:10.1016/j.ympev.2012.08.023.
- Capella-Gutiérrez S, Silla-Martínez JM, Gabaldón T. 2009. trimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics. 25(15):1972–1973. doi:10.1093/bioinformatics/btp348.
- Chen Y, Gan S, Shao L, Cheng C, Hao J. 2016a. The complete mitochondrial genome of the Pazala timur (Lepidoptera: Papilionidae: Papilioninae). Mitochondrial DNA A DNA Mapp Seq Anal. 27(1):533–534. doi:10.3109/19401736.2014.905843.
- Chen Y, Gan S, Wang Y, Wang Y, Zuo N, Hao J. 2016b. The complete mitochondrial genome of the Byasa alcinous (Lepidoptera: Papilionidae: Papilioninae). Mitochondrial DNA A DNA Mapp Seq Anal. 27(2):850–851. doi:10.3109/19401736.2014.919472.
- Chen X, Hao J. 2016. The complete mitochondrial genome of Graphium chironides (Lepidoptera: Papilionidae: Papilioninae). Mitochondrial DNA A DNA Mapp Seq Anal. 27(6):4049–4051. doi:10.3109/19401736.2014.1003838.
- Condamine FL, Nabholz B, Clamens AL, Dupuis JR, Sperling FAH. 2018. Mitochondrial phylogenomics, the origin of swallowtail butterflies, and the impact of the number of clocks in Bayesian molecular dating. Syst Entomol. 43(3):460–480. doi:10.1111/syen.12284.
- Cong Q, Borek D, Otwinowski Z, Grishin NV. 2015. Tiger swallowtail genome reveals mechanisms for speciation and caterpillar chemical defense. Cell Rep. 10(6):910–919. doi:10.1016/j.celrep.2015.01.026.
- Dong Y, Zhu L-X, Ding M-J, Wang J-J, Luo L-G, Liu Y, Ou Y-Y. 2016. Complete mitochondrial genome of Papilio syfanius (Lepidoptera: Papilionidae). Mitochondrial DNA A DNA Mapp Seq Anal. 27(1):403–404. doi:10.3109/19401736.2014.898278.
- Dong Y, Zhu L-X, Wu Y-F, Wu X-B. 2013. The complete mitochondrial genome of the Alpine black swallowtail, Papilio Maackii (Insecta: Lepidoptera: Papilionidae). Mitochondrial DNA. 24(6):639–641. doi:10.3109/19401736.2013.772162.
- Duan K, Zhang X, Zhang H-H, Hu S-J. 2020. Complete mitochondrial genome of the subalpine swordtail butterfly Graphium (Pazala) parus (Nicéville, 1900) (Lepidoptera: Papilionidae). Mitochondrial DNA Part B. 5(2):1903–1904. doi:10.1080/23802359.2020.1754945.
- He J-W, Zhang R, Yang J, Chang Z, Zhu L-X, Lu S-H, Xie F-A, Mao J-L, Dong Z-W, Liu G-C, et al. 2022. High-quality reference genomes of swallowtail butterflies provide insights into their coloration evolution. Zool Res. 43(3):367–379. doi:10.24272/j.issn.2095-8137.2021.303.
- Huang C-B, Zeng J-P, Zhou S-Y. 2015. Complete mitochondrial genomes of Teinopalpus imperialis (Lepidoptera: Papilionidae) and phylogenetic relationships analyses. Mitochondrial DNA A DNA Mapp Seq Anal. 27(4):2903–2904. doi:10.3109/19401736.2015.1060431.
- Jeng M-L, Chen M-Y, Wu L-W. 2021. Two complete mitochondrial genomes of Papilio butterflies obtained from historical specimens (Lepidoptera: Papilionidae). Mitochondrial DNA B Resour. 6(4):1341–1343. doi:10.1080/23802359.2021.1909443.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780. doi:10.1093/molbev/mst010.
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, et al. 2012. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 28(12):1647–1649. doi:10.1093/bioinformatics/bts199.
- Kong W, Hou Y, Zhang G, Yang T, Xiao R. 2019. Complete mitochondrial genome of Graphium doson (Papilioninae: Leptocircini). Mitochondrial DNA Part B. 4(1):698–699. doi:10.1080/23802359.2019.1574624.
- Kumar S, Stecher G, Li M, Knyaz C, Tamura K. 2018. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. 35(6):1547–1549. doi:10.1093/molbev/msy096.
- Lartillot N, Rodrigue N, Stubbs D, Richer J. 2013. PhyloBayes MPI: phylogenetic reconstruction with infinite mixtures of profiles in a parallel environment. Syst Biol. 62(4):611–615. doi:10.1093/sysbio/syt022.
- Laslett D, Canbäck B. 2008. ARWEN: a program to detect tRNA genes in metazoan mitochondrial nucleotide sequences. Bioinformatics. 24(2):172–175. doi:10.1093/bioinformatics/btm573.
- Liu G, Chang Z, Chen L, He J, Dong Z, Yang J, Lu S, Zhao R, Wan W, Ma G, et al. 2020. Genome size variation in butterflies (Insecta, Lepidoptera, Papilionoidea): a thorough phylogenetic comparison. Syst Entomol. 45(3):571–582. doi:10.1111/syen.12417.
- Miller JS. 1987. Phylogenetic studies in the Papilioninae (Lepidoptera: Papilionidae). Bull Am Mus Nat Hist. 186(4):365–512.
- Minh BQ, Schmidt HA, Chernomor O, Schrempf D, Woodhams MD, von Haeseler A, Lanfear R. 2020. IQ-TREE 2: new models and efficient methods for phylogenetic inference in the genomic era. Mol Biol Evol. 37(5):1530–1534. doi:10.1093/molbev/msaa015.
- Qin X-M, Guan Q-X, Li H-M, Zhang Y, Liu Y-J, Guo D-N. 2015. The complete mitogenome of Lamproptera curia (Lepidoptera: Papilionidae) and phylogenetic analyses of Lepidoptera. Front Biol. 10(5):458–472. doi:10.1007/s11515-015-1371-1.
- Qin F, Jiang G, Zhou S. 2012. Complete mitochondrial genome of the Teinopalpus aureus guangxiensis (Lepidoptera: Papilionidae) and related phylogenetic analyses. Mitochondrial DNA. 23(2):123–125. doi:10.3109/19401736.2011.653805.
- Shi M-R, Yu H, Xu J. 2020. The complete mitochondrial genome of the Papilio memno (Lepidoptera: Papilionidae). Mitochondrial DNA Part B. 5(3):2159–2160. doi:10.1080/23802359.2020.1768955.
- Simonsen TJ, Zakharov EV, Djernaes M, Cotton AM, Vane-Wright RI, Sperling FAH. 2011. Phylogenetics and divergence times of Papilioninae (Lepidoptera) with special reference to the enigmatic genera Teinopalpus and Meandrusa. Cladistics. 27(2):113–137. doi:10.1111/j.1096-0031.2010.00326.x.
- Sun Y, Huang H, Zhang X, Xia J, Geng J, Zhang K. 2020. The complete mitochondrial genome of the Papilio paris (Lepidoptera: Papilionidae). Mitochondrial DNA B Resour. 5(1):733–735. doi:10.1080/23802359.2020.1715281.
- Wu CS. 2001. Lepidopteran Papilionidae, class Insecta, Volume 25, zoology of China. Beijing: Science Press, p. 1–367.
- Wu L-W, Lees DC, Yen S-H, Hsu Y-F. 2010. The complete mitochondrial genome of the near-threatened swallowtail, Agehana maraho (Lepidoptera: Papilionidae): evaluating sequence variability and suitable markers for conservation genetic studies. Entomol News. 121(3):267–280. doi:10.3157/021.121.0308.
- Wu Y, Song J, Liu N, Cai Z, Yulin A, Fang J. 2016. The complete mitochondrial genome of Papilio xuthus (Lepidoptera: Papilionidae) and implications for Papilioninae taxonomy. Mitochondrial DNA B Resour. 1(1):144–145. doi:10.1080/23802359.2016.1144101.
- Yan Z, Lu M, Luo S, He S, Fu W, Wang X, Fan Z, Hu D, Chen B. 2022. Complete mitochondrial genomes of Papilio nephelus chaon and Papilio epycides (Lepidoptera: Papilionidae: Papilioninae) and phylogenetic analysis. Mitochondrial DNA B Resour. 7(7):1203–1205. doi:10.1080/23802359.2022.2091491.
- Zuo R, Jiang P, Sun C. 2018. The analysis of genetic variation in the mitochondrial genome and its application for the identification of Papilio species. Mitochondrial DNA Part B. 3(2):687–690.