Abstract
Meroplius fukuharai is an important sanitary and ecological resource insect. We sequenced and annotated the mitogenome of Meroplius fukuharai which is the first representative of the genus Meroplius with nearly complete mitochondrial data. This mitogenome is 14,803 bp long, which consists of 22 transfer RNA genes, 13 protein-coding genes (PCGs), and two ribosomal RNA genes. All genes have a conservational arrangement with other published species of Sepsidae. Our results also supported the monophyly of Sepsidae, and the genus Meroplius is more closely related to genus Sepsis, Microsepsis, and Archisepsis.
Introduction
Sepsidae is a globally distributed fly family with more than 320 described species (Ozerov Citation2005), and it is a relatively small, morphologically and ecologically uniform family of the Sciomyzoidea in the acalyptrate series of Diptera (Pont and Meier Citation2002). Meroplius fukuharai was originally described as the new species of the genus Xenosepsis (Iwasa Citation1984) and it was later transferred to the genus Meroplius based on morphology and molecular data (Meier Citation1995a, Citation1995b, Citation1996; Su et al. Citation2008; Zhao L et al. Citation2013). M. fukuharai flies mainly feed on pig dung and vertebrate carcasses (Iwasa Citation1984; Ozerov Citation1989; Ozerov Citation1991), and they can also spread diseases to cause myiasis (Xiao and Du Citation2006); therefore, M. fukuharai is an important sanitary and ecological resource insect. Due to their tiny size, usually 2–12 mm, it is very difficult to extract enough genomic DNA. Hence, we sequenced the mt genome of M. fukuharai, which is the first to be reported in the genus Meroplius. This study develops mt genome data of Sepsidae species, contributes to the discussion on the evolutionary relationships.
Materials and methods
The specimens of M. fukuharai () were collected on 18 July 2018 from Ivory Mountain (124.215613°E, 42.395751°N) Tieling City, Liaoning Province, China by Dan Yu. Each specimen was identified by Jianfeng Wang according to taxonomic keys (Pont and Meier Citation2002). The voucher specimens and their DNA were deposited in the Natural History Museum of Shenyang University (Jianfeng Wang, [email protected]) under the voucher number Sepsidae003.
Figure 1. Meroplius fukuharai (Iwasa). ♂ body in left lateral view. The photo was taken by Yingying Song on 7 September 2023.

The genomic DNA was extracted from the adult’s muscle tissues of the leg using the TIANamp Genomic DNA Kit (TIANGEN, Beijing, China), and sequenced under the first generation sequencing technology with 22 primer pairs (Table S1; Supplementary Information) (Simon et al. Citation1994; Weigl et al. Citation2010; Zhang NX et al. Citation2013; Zhao Z et al. Citation2013). DNA fragments were sequenced on both strands by Applied Biosystems 3730 DNA Analyzer (Sangon Biotech Co. Ltd., Shanghai, China). Genome sequence was assembled by DNAman Version 6 software and was annotated by MITOS2 online software (http://mitos2.bioinf.uni-leipzig.de). The annotated results were confirmed by BLAST in GenBank with Nemopoda mamaevi (KM605250). A total of 15 species (Table S2) were used in Bayesian inference (BI) phylogenetic analysis based on 13 PCGs by PhyloSuite (Zhang et al. Citation2020). The Best-fit model according to BIC: GTR + F + I + G4.
Results
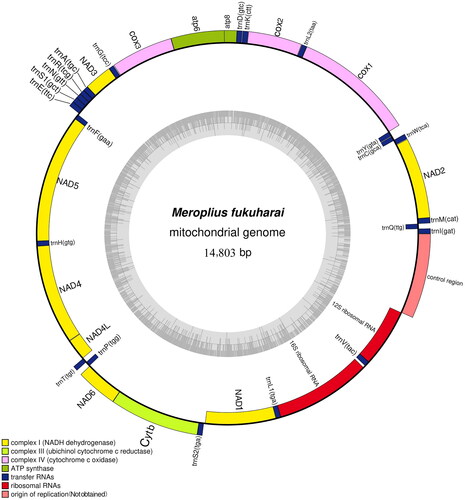
This mitogenome of M. fukuharai is 14,803 bp long, which contains 22 transfer RNA genes, 13 protein-coding genes (PCGs), and two ribosomal RNA genes (). The nucleotide composition of the mitogenome was 37.41% of A, 36.63% of T, 15.90% of C, and 10.07% of G. The A + T content was 74.04% which conforms to the characteristics of AT preference like other flies. The codon ATG was the most popular start codon shared with the COX2, ATP6, COX3, NAD4, NAD4L, Cytb, and start codon ATT was shared with the NAD2, NAD3, NAD5, and NAD6. Particularly, the COX1 begins with codon TCG, the ATP8 begins with codon ATA and the NAD1 begins with codon TTG. The conservative stop codon TAA was shared with the NAD2, COX1, COX2, ATP8, ATP6, COX3, NAD4L, and NAD6. Another common stop codon TAG was shared with NAD3, Cytb, and NAD1. Particularly, the NAD5 and NAD4 stop with codon T. The two rRNA genes (16S rRNA and 12S rRNA) are separated by trnV(tac). The longest gene spacer is 29 bp, located between 16S rRNA and trnV(tac), and the longest gene overlap is 23 bp, located between trnL1(tga) and 16S rRNA. The 22 tRNA genes are interspersed between rRNAs and PCGs, with sizes ranging from 63 bp (trnR(tcg)) to 77 bp (trnV(tac)).
Figure 2. Gene map of the mitochondrial genome of Meroplius fukuharai (GenBank accession number: OR270120), with 13 protein-coding genes, 22 tRNAs, and two rRNAs.
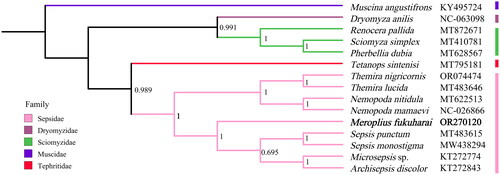
The topology of BI phylogenetic tree is shown in . According to the phylogenetic outcome, the outgroup Muscina angustifrons of Muscidae is a single branch, three Sciomyzoidea species and one Dryomyzidae species form a clade, and nice Sepsidae species form a clade. Meroplius fukuharai, two species of Sepsis, Microsepsis sp., and Archisepsis discolor cluster together with strong support.
Figure 3. Bayesian phylogenetic tree of 15 Diptera species. The posterior probabilities are labeled at each node (0.6 and greater). GenBank accession numbers of all sequence used in the phylogenetic tree have been included in figure and corresponding to the names of all species. The following sequences were used: Archisepsis discolor (KT272843, Junqueira et al. Citation2016), Dryomyza anilis (NC_063098), Meroplius fukuharai (OR270120), Microsepsis sp. (KT272774, Junqueira et al. Citation2016), Muscina angustifrons (KY495724, Karagozlu et al. Citation2017), Nemopoda mamaevi (NC_026866, Li et al. Citation2015), Nemopoda nitidula (MT622513), Pherbellia dubia (MT628567), Renocera pallida (MT872671), Sciomyza simplex (MT410781), Sepsis monostigma (MW438294, Yang and Wang Citation2021), Sepsis punctum (MT483615), Tetanops sintenisi (MT795181), Themira lucida (MT483646), and Themira nigricornis (OR074474, Wong et al. Citation2023).
Discussion and conclusions
In this study, the mitogenome of M. fukuharai was sequenced and annotated for the first time. It was similar with related species reported before (Simon et al. Citation1994; Zhang NX et al. Citation2013; Zhao L et al. Citation2013; Zhao Z et al. Citation2013; Li et al. Citation2015; Chen et al. Citation2018; She et al. Citation2020; Yang and Wang Citation2021). Our results supported the monophyly of Sepsidae, and this result is the same as previous studies based on morphology and molecular data (Meier Citation1995a, Citation1995b, Citation1996; Su et al. Citation2008; Zhao L et al. Citation2013; Zhao Z et al. Citation2013). The genus Meroplius is more closely related to genus Sepsis, Microsepsis, and Archisepsis. The mt genome of M. fukuharai could provide important information for further studies of Sepsidae phylogeny.
Author contributions
Jianfeng Wang designed the research study and obtained the funding. Yingying Song performed laboratory work. Yingying Song analyzed and interpreted the data for the work. Jianfeng Wang and Yingying Song wrote and revised the manuscript. All authors read and approved the final manuscript before submission.
Ethical approval
This study was approved by the Science and Technology Ethics Committee of Shenyang University.
Supplemental Material
Download PNG Image (238.1 KB)Supplemental Material
Download PNG Image (15.2 KB)Supplemental Material
Download PNG Image (250.6 KB)Supplemental Material
Download MS Word (525.1 KB)Supplemental Material
Download MS Word (15.2 KB)Supplemental Material
Download MS Word (14.1 KB)Supplemental Material
Download PNG Image (90.4 KB)Supplemental Material
Download PDF (168.7 KB)Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI at https://www.ncbi.nlm.nih.gov/ under the accession no. OR270120.
Additional information
Funding
References
- Chen W, Shang YJ, Ren LP, Zhang XY, Guo YD. 2018. The complete mitochondrial genome of Graphomya rufitibia (Diptera: Muscidae). Mitochondrial DNA B Resour. 3(1):403–404. doi:10.1080/23802359.2018.1456375.
- Iwasa M. 1984. Studies on the Sepsidae from Japan (Diptera). III. On the eleven species of eight genera excluding the genera Sepsis Fallén and Themira R.-D., with description of a new species. Kontyû. 52(2):296–308.
- Junqueira AC, Azeredo-Espin AM, Paulo DF, Marinho MA, Tomsho LP, Drautz-Moses DI, Purbojati RW, Ratan A, Schuster SC. 2016. Large-scale mitogenomics enables insights into Schizophora (Diptera) radiation and population diversity. Sci Rep. 6(1):21762. doi:10.1038/srep21762.
- Karagozlu MZ, Park SH, Shin S-E, Kim C-B. 2017. Sequencing and analyzing complete mitochondrial DNA of Muscina angustifrons (Insecta, Diptera, Muscidae). Mitochondrial DNA B Resour. 2(1):115–116. doi:10.1080/23802359.2017.1289352.
- Li XK, Ding SM, Cameron SL, Kang ZH, Wang YY, Yang D. 2015. The first mitochondrial genome of the sepsid fly Nemopoda mamaevi Ozerov, 1997 (Diptera: Sciomyzoidea: Sepsidae), with mitochondrial genome phylogeny of Cyclorrhapha. Public Libr Sci. 10(3):e0123594.
- Meier R. 1995a. Cladistic analysis of the Sepsidae (Cyclorrhapha: Diptera) based on a comparative scanning electron microscopic study of larvae. Syst Entomol. 20(2):99–128. doi:10.1111/j.1365-3113.1995.tb00086.x.
- Meier R. 1995b. A comparative SEM study of the eggs of the Sepsidae (Diptera) with a cladistic analysis based on egg, larval and adult characters. Insect Syst Evol. 26(4):425–438. doi:10.1163/187631295X00080.
- Meier R. 1996. Larval morphology of the Sepsidae (Diptera, Sciomyzoidea), with a cladistic analysis using adult and larval characters. Bull Am Mus Nat Hist. 228:1–147.
- Ozerov AL. 1989. Flies of the families Sepsidae and Piophilidae (Diptera) of the Zejskij State Nature Preserve. Entomol Oboz. 68(4):839–849.
- Ozerov AL. 1991. Biology and morphology of the larvae of Palaearctic species of the genera Meroplius R.-D. and Xenosepsis Malloch (Diptera, Sepsidae). Biol Nauki. 1:49–54.
- Ozerov AL. 2005. World catalogue of the family Sepsidae (Insecta: Diptera). Zool Stud. 8:1–74.
- Pont AC, Meier R. 2002. The Sepsidae (Diptera) of Europe. Fauna Entomol Scand. 37:1–221.
- She YT, Wang CJ, Wang WF, Zeng Q, Mao W, Gu XP, Yuan H, Pan XL, Wang Y. 2020. The whole mitochondrial genome of Sarcophaga kanoi (Diptera: Sarcophagidae). Mitochondrial DNA B Resour. 5(3):2648–2649. doi:10.1080/23802359.2020.1787272.
- Simon C, Frati F, Beckenbach A, Crespi B, Liu H, Flook P. 1994. Evolution, weighting, and phylogenetic utility of mitochondrial gene sequences and a compilation of conserved polymerase chain reaction primers. Ann Entomol Soc Am. 87(6):651–701. doi:10.1093/aesa/87.6.651.
- Su KFY, Kutty SN, Meier R. 2008. Morphology versus molecules: the phylogenetic relationships of Sepsidae (Diptera: Cyclorrhapha) based on morphology and DNA sequence data from ten genes. Cladistics. 24(6):902–916. doi:10.1111/j.1096-0031.2008.00222.x.
- Weigl S, Testini G, Parisi A, Dantas-Torres F, Traversa D, Colwell DD, Otranto D. 2010. The mitochondrial genome of the common cattle grub, Hypoderma lineatum. Med Vet Entomol. 24(3):329–335. doi:10.1111/j.1365-2915.2010.00873.x.
- Wong D, Norman H, Creedy TJ, Jordaens K, Moran KM, Young A, Mengual X, Skevington JH, Vogler AP. 2023. The phylogeny and evolutionary ecology of hoverflies (Diptera: Syrphidae) inferred from mitochondrial genomes. Mol Phylogenet Evol. 184:107759. doi:10.1016/j.ympev.2023.107759.
- Xiao CX, Du YZ. 2006. Advance in researches on the systematics of Sepsidae (Diptera: Sciomyzoidea). Entomol J East China. 15(4):244–249.
- Yang P, Wang JF. 2021. The first mitochondrial genome of Sepsis monostigma (Diptera: Sepsidae). Mitochondrial DNA B Resour. 6(9):2517–2518. doi:10.1080/23802359.2021.1959431.
- Zhang D, Gao F, Jakovlić I, Zou H, Zhang J, Li WX, Wang GT. 2020. PhyloSuite: an integrated and scalable desktop platform for streamlined molecular sequence data management and evolutionary phylogenetics studies. Mol Ecol Resour. 20(1):348–355. doi:10.1111/1755-0998.13096.
- Zhang NX, Zhang YJ, Yu G, Chen B. 2013. Structure characteristics of the mitochondrial genomes of Diptera and design and application of universal primers for their sequencing. Acta Parasitol Med Entomol Sin. 56(4):398–407.
- Zhao L, Annie ASH, Amrita S, Yi SKF, Rudolf M. 2013. Does better taxon sampling help? A new phylogenetic hypothesis for Sepsidae (Diptera: Cyclorrhapha) based on 50 new taxa and the same old mitochondrial and nuclear markers. Mol Phylogenet Evol. 69(1):153–164. doi:10.1016/j.ympev.2013.05.011.
- Zhao Z, Su TJ, Chesters D, Wang SD, Ho SYW, Zhu CD, Chen XL, Zhang CT. 2013. The mitochondrial genome of Elodia flavipalpis Aldrich (Diptera: Tachinidae) and the evolutionary timescale of Tachinid flies. PLOS One. 8(4):e61814. doi:10.1371/journal.pone.0061814.
