Abstract
Sucra jujuba Chu, Citation1979 (Lepidoptera: Geometridae) is a major insect pest in jujube plantation. In this study, we have sequenced the complete mitochondrial genome of S. jujuba. The circular genome was 15,557 bp in length and contained 13 protein-coding genes (PCGs), 22 transfer RNA genes (tRNAs), two ribosomal RNA genes (rRNAs), and one AT-rich region (GenBank accession no. MZ507574). The nucleotide composition was significantly biased (A, T, C, and G were 41.85%, 39.65%, 10.97%, and 7.53%, respectively) with A + T contents of 81.50%. The Bayesian phylogenetic analysis of the concatenated nucleotide sequences of 13 PCGs from 30 species in the subfamily Ennominae and two outgroup species was performed. The results indicated that S. jujuba was closely related to Amraica recursaria in the subfamily Ennominae.
Introduction
The jujube geometrid, Sucra jujuba Chu, Citation1979 is native to northern regions of China where it has become a major leaf-feeding pest of Chinese jujube trees (Ziziphus jujuba Mill.) (Chu Citation1979). The male moth has pectinate antennae and a gray-brown body, and its body length is 12–13 mm. It has a wing span of about 35 mm and is characterized by a noticeable black corrugated line in the middle of hindwings, while the female moth has filiform antennae, a body length of about 15 mm and is wingless () (Li et al. Citation1981; Li Citation1992). The larvae of S. jujuba feed solely on the leaves of jujube trees and seriously affect the yield and quality of jujube. It is widely distributed in Northern China (Henan, Hebei, Shandong, Shanxi, Shaanxi, and Ningxia Provinces) and has caused considerable economic losses to jujube industry (Hou et al. Citation2002; Li et al. Citation2016). In this study, we sequenced and assembled the complete mitogenome of Sucra jujuba, and performed the phylogenetic analysis among the species in the subfamily Ennominae, which will contribute to get a better understanding of the phylogenetic relationships of Sucra jujuba and provide new ideas for jujube pest control.
Figure 1. Female specimen of Sucra jujuba Chu, Citation1979 was collected from a jujube orchard in Shenmu County, Yulin City, China. Photographed by Bo Hong on 17 June 2020.

Materials and methods
Sample collection and genomic DNA extraction
The female specimen of S. jujuba was collected from a jujube orchard in Shenmu County, Yulin City, Shaanxi Province, China (110.5663° E, 38.2885° N, altitude 716.1 m). The genomic DNA was extracted from the muscle tissues of the specimen’s thorax using TIANamp Genomic DNA Kit (Tiangen, Beijing, China). The quality and quantity of the DNA were assessed by 1.0% agarose gel electrophoresis and SimpliNano spectrophotometer (GE Healthcare, Piscataway, NJ). The specimen was deposited in the Laboratory of Pest Monitoring and Control Center, Bio-Agriculture Institute of Shaanxi, Xi’an, Shaanxi, China (http://swny.ac.cn/, contact person and email: Bo Hong, [email protected]) under the voucher number SN2020ZCH01.
Mitochondrial genome sequencing, assembly, and annotation
The complete mitochondrial genome sequencing was performed on Illumina NovaSeq 6000 platform with 150 bp paired-end reads at Biomarker Technologies Co., Ltd. (Beijing, China). The mitogenome sequence was assembled using MITObim v1.9.1 (Hahn et al. Citation2013) with the species Ectropis grisescens (GenBank accession no. MW337302) (Song et al. Citation2021) as the reference genome. The average coverage depth for the mitochondrial genome sequence was calculated using Bowtie2 v 2.3.4 (Langmead and Salzberg Citation2012) and SAMtools v1.7 (https://github.com/samtools/samtools; Danecek et al. Citation2021) to confirm the assembly correctness. The rough boundaries of each gene were first identified using the MITOS2 Web Server (http://mitos2.bioinf.uni-leipzig.de/index.py; Bernt et al. Citation2013) with the invertebrate genetic code. The nucleotide sequences of 13 PCGs and two rRNA genes were further confirmed by the published lepidopteran mitogenomes using MEGA 11 (Tamura et al. Citation2021). The positions of tRNA genes were determined using the tRNAscan-SE 2.0 Search Server (http://lowelab.ucsc.edu/tRNAscan-SE/; Chan et al. Citation2021). Finally, the circular genome visualization was generated using Proksee (https://proksee.ca/; Grant et al. Citation2023). To better understand the base usage bias of the mitogenome, we also calculated the nucleotide skew values using the formulae AT-skew = (A − T)/(A + T) and GC-skew = (G − C)/(G + C) (Perna and Kocher Citation1995).
Phylogenetic analysis
Phylogenetic analysis was conducted using the mitogenome sequences of 30 species (including S. jujuba) in the subfamily Ennominae and two outgroup species in the family Geometridae, Idaea effusaria Christoph, 1881 (Sterrhinae) and Pasiphila chloerata Mabille, 1870 (Larentiinae). The sequences were downloaded from the GenBank database (Wang et al. Citation2017; Chen et al. Citation2019; Du et al. Citation2019; Song et al. Citation2019; Xie Citation2020; Huang et al. Citation2021; Song et al. Citation2021; Sun et al. Citation2021; Chen et al. Citation2022; Lu et al. Citation2023; Zou et al. Citation2023). All nucleotide sequences of 13 protein-coding genes (PCGs) for these species were aligned using MAFFT7.037 with L-INS-i strategy (Katoh et al. Citation2002) and concatenated using DAMBE 6.4.81 (Xia Citation2017). The Bayesian inference (BI) phylogenetic tree was constructed using MrBayes3.2.6 (Ronquist and Huelsenbeck Citation2003). Two independent runs with four Markov chains were performed for 1,000,000 generations and sampled every 1000 generations. Finally, the phylogenetic tree was visualized and edited with FigTree 1.4.4 (http://tree.bio.ed.ac.uk/software/figtree/).
Results
Characteristics of mitochondrial genome
The complete mitochondrial genome of Sucra jujuba (GenBank accession no. MZ507574) is a circular molecule of 15,557 bp in length, and consists of 37 genes, including 13 PCGs, 22 transfer RNA genes (tRNAs), two ribosomal RNA genes (rRNAs), and one control region (non-coding AT-rich region) (). The average coverage depth of the mitochondrial genome is 2762.45× (Supplementary Figure S1). The nucleotide composition is significantly biased (A, T, C, and G are 41.85%, 39.65%, 10.97%, and 7.53%, respectively) with A + T contents of 81.50%. AT-skew and GC-skew are 0.027 and −0.186, respectively. There are 17 intergenic spacer regions ranging in size from 1 to 66 bp (255 bp in total) and five overlapping regions (24 bp in total) throughout the whole genome.
Figure 2. The mitochondrial genome map of Sucra jujuba Chu, Citation1979 with 13 PCGs, 22 tRNAs, two rRNAs, and one at-rich region.
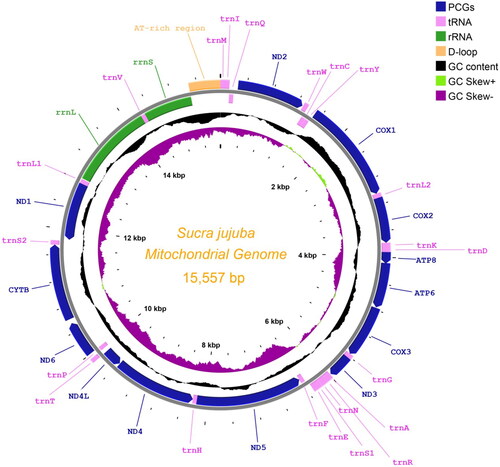
Among 13 PCGs, except for cox1 which uses CGA as an initiation codon, all other PCGs are initiated with typical ATN codons (nad2, nad3, and nad5 with ATT, nad6 and nad1 with ATA, cox2, atp6, cox3, nad4, nad4l, and cytb with ATG, and atp8 with ATC). All the PCGs have a typical TAN stop codon; nine PCGs (nad2, atp8, atp6, cox3, nad3, nad4l, nad6, cytb, and nad1) are terminated with TAA, and four PCGs (cox1, cox2, nad4, and nad5) are terminated with an incomplete stop codon T. All 22 tRNAs range from 65 to 71 bp, and are predicted to contain typical cloverleaf secondary structures except the gene trnS1, whose DHU arm is replaced by a simple loop. The two rRNA genes are encoded on the N-strand of the mitochondrial genome. rrnL (16S rRNA) is 1377 bp in length with A + T contents of 84.75%, and rrnS (12S rRNA) is 784 bp in length with A + T contents of 85.71%. The AT-rich region is 473 bp in length with A + T contents of 95.98%, and is located between trnM and rrnS ().
Phylogenetic analysis
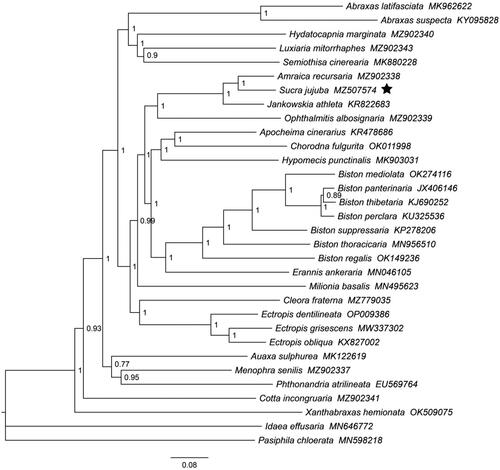
Phylogenetic analysis of S. jujuba, along with 29 species in the subfamily Ennominae, was performed using the BI method, with I. effusaria and P. chloerata being used as outgroup species. The phylogenetic tree showed that Sucra jujuba was more closely related to Amraica recursaria than to other species with high Bayesian posterior probability (BPP = 1) ().
Figure 3. The Bayesian phylogenetic tree of S. jujuba with 29 species in the subfamily Ennominae based on the concatenated nucleotide sequences of 13 PCGs. The black pentagram represented the branch of S. jujuba. The sequences used to construct the tree were as follows: Abraxas latifasciata MK962622, Abraxas suspecta KY095828 (Sun et al. Citation2017), Hydatocapnia marginata MZ902340, Luxiaria mitorrhaphes MZ902343, Semiothisa cinerearia MK880228 (Zou et al. Citation2023), Amraica recursaria MZ902338, Sucra jujuba MZ507574, Jankowskia athlete KR822683 (Xu et al. Citation2016), Ophthalmitis albosignaria MZ902339 (Zheng et al. Citation2022), Apocheima cinerarius KR478686 (Liu et al. Citation2014), Chorodna fulgurita OK011998, Hypomecis punctinalis MK903031 (Sun et al. Citation2021), Biston mediolata OK274116, Biston panterinaria JX406146 (Yang et al. Citation2013), Biston thibetaria KJ690252 (Chen et al. Citation2017), Biston perclara KU325536 (Chen et al. Citation2017), Biston suppressaria KP278206 (Chen et al. Citation2016), Biston thoracicaria MN956510 (Huang et al. Citation2021), Biston regalis OK149236, Erannis ankeraria MN046105 (Chen et al. Citation2019), Milionia basalis MN495623 (Du et al. Citation2019), Cleora fraternal MZ779035, Ectropis dentilineata OP009386 (Lu et al. Citation2023), Ectropis grisescens MW337302 (Song et al. Citation2021), Ectropis obliqua KX827002 (Wang et al. Citation2017), Auaxa sulphurea MK122619, Menophra senilis MZ902337, Phthonandria atrilineata EU569764 (Yang et al. Citation2009), Cotta incongruaria MZ902341, Xanthabraxas hemionata OK509075 (Chen et al. Citation2022), Idaea effusaria MN646772 (Xie Citation2020), and Pasiphila chloerata MN598218 (Song et al. Citation2019). Bayesian posterior probabilities were shown for each node.
Discussion and conclusions
In this study, we assembled and described the characteristics of the mitochondrial genome of S. jujuba. The gene arrangement of the mitogenome of S. jujuba is identical to that of most of lepidopteran mitochondrial genomes with the ‘trnM-trnI-trnQ’ cluster (Wang et al. Citation2017; Huang et al. Citation2021; Zou et al. Citation2023). The phylogenetic analysis indicated that S. jujuba was more closely related to A. recursaria than to other species in the subfamily Ennominae, which was consistent with the previous study by Zou et al. (Citation2023). This study offers significant molecular data for advancing the evolutionary and phylogeographic analysis of S. jujuba, as well as establishing a foundation for further investigation into the phylogenetic relationships among species in the subfamily Ennominae.
Author contributions
X.L.W. and B.H. contributed to the conception and design of the research. Y.Y.Z., K.W., and B.H. collected the specimens and performed the experiments. X.L.W., Y.L., and B.H. analyzed the data. X.L.W. and Y.Y.Z. drafted the manuscript, Y.L. and B.H. revised the manuscript. All authors approved the final version of the manuscript and agreed to be accountable for all aspects of the work.
Ethics statement
There is no ethical research involved in this study. The specimen collection protocol was approved by Shaanxi Academy of Sciences. The study did not involve endangered or protected species.
Supplemental Material
Download MS Word (167.5 KB)Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI at https://www.ncbi.nlm.nih.gov/ under the accession no. MZ507574. The associated BioProject, Bio-Sample, and SRA numbers are PRJNA758210, SAMN21016412, and SRR15650334, respectively.
Additional information
Funding
References
- Bernt M, Donath A, Jühling F, Externbrink F, Florentz C, Fritzsch G, Pütz J, Middendorf M, Stadler PF. 2013. MITOS: improved de novo metazoan mitochondrial genome annotation. Mol Phylogenet Evol. 69(2):313–319. doi:10.1016/j.ympev.2012.08.023.
- Chan PP, Lin BY, Mak AJ, Lowe TM. 2021. tRNAscan-SE 2.0: improved detection and functional classification of transfer RNA genes. Nucleic Acids Res. 49(16):9077–9096. doi:10.1093/nar/gkab688.
- Chen HY, Li W, Shen S. 2022. Characterization and phylogenetic analysis of the mitochondrial genome of Xanthabraxas hemionata (Lepidoptera: Geometridae). Mitochondrial DNA B Resour. 7(8):1481–1483. doi:10.1080/23802359.2022.2107447.
- Chen R, Xue D, Galsworthy A, Han H. 2017. Complete mitochondrial genomes throw light on budding speciation in three Biston species (Lepidoptera, Geometridae). Zool Scripta. 46(1):73–84. doi:10.1111/zsc.12184.
- Chen SC, Wang XQ, Wang JJ, Hu X, Peng P. 2016. The complete mitochondrial genome of a tea pest looper, Buzura suppressaria (Lepidoptera: Geometridae). Mitochondrial DNA A DNA Mapp Seq Anal. 27(5):3153–3154. doi:10.3109/19401736.2015.1007310.
- Chen YM, Wang QH, Wang SB, Qu LJ. 2019. The mitochondrial genome of Erannis ankeraria (Lepidoptera: Geometridae). Mitochondrial DNA B Resour. 4(2):2696–2697. doi:10.1080/23802359.2019.1644560.
- Chu HF. 1979. Three new genera and three new species of the family Geometridae. Acta Entomol Sin. 22(1):73–76. doi:10.16380/j.kcxb.1979.01.009.
- Danecek P, Bonfield JK, Liddle J, Marshall J, Ohan V, Pollard MO, Whitwham A, Keane T, McCarthy SA, Davies RM, et al. 2021. Twelve years of SAMtools and BCFtools. Gigascience. 10(2):giab008. doi:10.1093/gigascience/giab008.
- Du YM, Song X, Lu ZJ. 2019. Phylogenetic relationship and characterization of the complete mitochondrial genome of Milionia basalis (Lepidoptera: Geometridae). Mitochondrial DNA B Resour. 4(2):4136–4137. doi:10.1080/23802359.2019.1692732.
- Grant JR, Enns E, Marinier E, Mandal A, Herman EK, Chen C, Graham M, Van Domselaar G, Stothard P. 2023. Proksee: in-depth characterization and visualization of bacterial genomes. Nucleic Acids Res. 51(W1):W484–W492. doi:10.1093/nar/gkad326.
- Hahn C, Bachmann L, Chevreux B. 2013. Reconstructing mitochondrial genomes directly from genomic next-generation sequencing reads—a baiting and iterative mapping approach. Nucleic Acids Res. 41(13):e129. doi:10.1093/nar/gkt371.
- Hou BL, Zhao JW, Han DD, Sun XH, Dong ZB, Zhu Q. 2002. New research advances on Sucra jujuba. Shandong For Sci Technol. 3:35–38. doi:10.3969/j.issn.1002-2724.2002.03.020.
- Huang BS, Zhao YJ, Wu YP. 2021. Complete mitochondrial genome of Biston thoracicaria (Lepidoptera: Geometridae). Mitochondrial DNA B Resour. 6(7):2007–2008. doi:10.1080/23802359.2021.1939181.
- Katoh K, Misawa K, Kuma K, Miyata T. 2002. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 30(14):3059–3066. doi:10.1093/nar/gkf436.
- Langmead B, Salzberg SL. 2012. Fast gapped-read alignment with Bowtie 2. Nat Methods. 9(4):357–359. doi:10.1038/nmeth.1923.
- Li LC, Dong HF, Yan ZQ. 1981. Occurrence and integrated control technology for Sucra jujuba. J Shanxi Agric Univ. 1:69–84. doi:10.13842/j.cnki.issn1671-8151.1981.01.007.
- Li LC. 1992. Chinese jujube pest. Beijing: China Agriculture Press.
- Li ZW, Li P, Wang DJ, Ji XS. 2016. Integration and application of integrated prevention and control for Sucra jujuba in Ningxia jujube plantation. Ningxia Agric For Sci Technol. 57(9):32–34. doi:10.3969/j.issn.1002-204X.2016.09.013.
- Liu S, Xue D, Cheng R, Han H. 2014. The complete mitogenome of Apocheima cinerarius (Lepidoptera: Geometridae: Ennominae) and comparison with that of other lepidopteran insects. Gene. 547(1):136–144. doi:10.1016/j.gene.2014.06.044.
- Lu Z, Yuan R, Tang P, Wang Z, Zhou X, Chen X. 2023. The mitochondrial genome of Ectropis dentilineata Moore, 1868 (Lepidoptera: Geometridae) from tea plantations in Guizhou Province, China. All Life. 16(1):2246665. doi:10.1080/26895293.2023.2246665.
- Perna NT, Kocher TD. 1995. Patterns of nucleotide composition at fourfold degenerate sites of animal mitochondrial genomes. J Mol Evol. 41(3):353–358. doi:10.1007/BF01215182.
- Ronquist F, Huelsenbeck JP. 2003. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 19(12):1572–1574. doi:10.1093/bioinformatics/btg180.
- Song L, Shi YX, Li JH, Zhang HF, Ding WL, Li J, Yang MS. 2019. Complete mitochondrial genome of the Pasiphila chloerata (Lepidoptera: Geometridae) and its phylogenetic implications. Mitochondrial DNA B Resour. 4(2):4142–4143. doi:10.1080/23802359.2019.1692725.
- Song XH, Yang TB, Xu XQ, Yan XH, Zhou CQ. 2021. Characterization of the complete mitochondrial genome of Ectropis grisescens (Lepidoptera: Geometridae). Mitochondrial DNA B Resour. 6(7):1953–1955. doi:10.1080/23802359.2021.1923423.
- Sun Y, Wang J, Li F, Huang X, Geng X, Li J. 2021. Complete mitochondrial genome of Hypomecis punctinalis Scopoli, 1763 and its phylogenetic position within family Geometridae. Mitochondrial DNA B Resour. 6(10):2910–2912. doi:10.1080/23802359.2021.1969698.
- Sun Y, Zhang J, Li Q, Liang D, Abbas MN, Qian C, Wang L, Wei G, Zhu BJ, Liu CL. 2017. Mitochondrial genome of Abraxas suspecta (Lepidoptera: Geometridae) and comparative analysis with other Lepidopterans. Zootaxa. 4254(5):501–519. doi:10.11646/zootaxa.4254.5.1.
- Tamura K, Stecher G, Kumar S. 2021. MEGA11: molecular evolutionary genetics analysis version 11. Mol Biol Evol. 38(7):3022–3027. doi:10.1093/molbev/msab120.
- Wang XQ, Chen SC, Li PW, Hu X, Peng P. 2017. The complete mitochondrial genome of a tea geometrid, Ectropis obliqua (Lepidoptera: Geometridae). Mitochondrial DNA B Resour. 2(2):459–460. doi:10.1080/23802359.2017.1357449.
- Xia X. 2017. DAMBE6: new tools for microbial genomics, phylogenetics and molecular evolution. J Hered. 108(4):431–437. doi:10.1093/jhered/esx033.
- Xie JL. 2020. Sequencing and characterization of mitochondrial genome of Idaea effusaria (Lepidoptera: Geometridae). Mitochondrial DNA B Resour. 5(1):1001–1002. doi:10.1080/23802359.2020.1720542.
- Xu YM, Chen SC, Wang XQ, Peng P, Li PW. 2016. The complete mitogenome of Jankowskia athleta (Lepidoptera: Geometridae). Mitochondrial DNA A DNA Mapp Seq Anal. 27(4):3035–3036. doi:10.3109/19401736.2015.1063122.
- Yang L, Wei ZJ, Hong GY, Jiang ST, Wen LP. 2009. The complete nucleotide sequence of the mitochondrial genome of Phthonandria atrilineata (Lepidoptera: Geometridae). Mol Biol Rep. 36(6):1441–1449. doi:10.1007/s11033-008-9334-0.
- Yang X, Xue D, Han H. 2013. The complete mitochondrial genome of Biston panterinaria (Lepidoptera: Geometridae), with phylogenetic utility of mitochondrial genome in the Lepidoptera. Gene. 515(2):349–358. doi:10.1016/j.gene.2012.11.031.
- Zheng X, Zhang R, Yue B, Wu Y, Yang N, Zhou C. 2022. Enhanced resolution of evolution and phylogeny of the moths inferred from nineteen mitochondrial genomes. Genes. 13(9):1634. doi:10.3390/genes13091634.
- Zou D, Yuan J, Ding J, Li J, Zhang H. 2023. The complete mitochondrial genome of Semiothisa cinerearia Bremer & Grey 1853 (Lepidoptera: Geometridae: Ennominae) and its phylogenetic analysis. Mitochondrial DNA B Resour. 8(11):1248–1252. doi:10.1080/23802359.2023.2278820.
