Abstract
Holocephali is a subclass of chondrichthyans with ample geographic distribution in marine ecosystems. Holocephalan species are organized into three families: Callorhinchidae, Chimaeridae, and Rhinochimaeridae. Despite the critical ecological and evolutionary importance, genomic information from holocephalans is still scarce, particularly from rhinochimaerids. The present study provides the first complete mitogenome of the Atlantic longnose chimaera Rhinochimaera atlantica (Holt & Byrne, 1909). The whole mitogenome was sequenced from an R. atlantica specimen, collected on the Porcupine Bank (NE Atlantic), by Illumina high-throughput sequencing. The R. atlantica mitogenome has 17,852 nucleotides with 13 protein-coding genes, 22 transfer RNA, and two ribosomal RNA genes. Nine of these genes are in the complementary strand. This mitogenome has a GC content of 41.5% and an AT content of 58.5%. The phylogenetic reconstruction provided here, using all the available complete and partial Holocephali mitogenomes, places R. atlantica in the Rhinochimaeridae family, as expected. This genomic resource will be useful in the genomic characterization of this species.
Introduction
Holocephali comprises mostly deep-water species widely distributed, except for the Antarctic ecosystem (Didier et al. Citation2012). This subclass of Chondrichthyes comprises nearly 40 species and only one taxonomic order (Chimaeriformes) with three families (Callorhinchidae, Chimaeridae, and Rhinochimaeridae) (Compagno et al. Citation2005; Weigmann Citation2016). Rhinochimaeridae specimens are characterized by a conspicuously long and flat snout, large head, and elongated body (Carpenter Citation2002; Didier et al. Citation2012) and inhabiting water depths between 400 m and more than 2000 m (Carpenter Citation2002; Weigmann Citation2016). Currently, this family comprises three genera (Harriotta, Neoharriotta, and Rhinochimaera) (Finucci et al. Citation2017). For the three Rhinochimaera species, two complete mitogenomes of Rhinochimaera pacifica and one partial mitogenome of Rhinochimaera africana are available (accessed on 1 December 2023). However, a complete mitogenome of Rhinochimaera atlantica (Holt & Byrne, 1909) (Atlantic longnose chimaera) is not available (accessed on 1 December 2023). The presence of R. atlantica has been documented in South Africa, the North Atlantic Ocean, the Gulf of Mexico (WORMS, accessed on 1 December 2023) (Carpenter Citation1969), and the western Indian Ocean (Weigmann Citation2016) (Supplementary Figure S1). Rhinochimaera atlantica is listed as ‘Least Concern’ by the International Union for Conservation of Nature (IUCN) (Finucci Citation2020). Yet, its biological characteristics as slow growth, low fecundity, or the tendency to aggregate contribute to its overfishing and consequent vulnerability (Stevens et al. Citation2000; Barnett et al. Citation2012; Finucci et al. Citation2017). The biological and evolutionary importance of Chondrichthyes has boosted the development of studies aiming to provide genomic resources helpful in the study of these species (e.g. Vilas-Arrondo et al. Citation2022; Yamaguchi et al. Citation2023). However, there is a substantial lack of molecular data which, in combination with morphological characters, is crucial for describing new Chimaeridae species and elucidating relationships among extant holocephalans (Licht et al. Citation2012; Iglésias et al. Citation2022). Hence, the present study provides the first complete mitogenome of R. atlantica.
Materials and methods
On 28 September 2021, a male specimen of R. atlantica was collected in the Porcupine Bank (51.301200, −13.211200)), North-East Atlantic, at a depth of 1435 m (Supplementary Figure S1) during the Spanish Bottom Trawl Survey on the Porcupine Bank. Sampling was carried out onboard the R/V Vizconde de Eza, a stern trawler of 53 m and 1800 kW, using a Baca-GAV 39/52 with a cod-end mesh size of 20 mm. The specimen (voucher name k141_3), which was morphologically identified on board, had a Pre Supra Caudal Fin Length (PSCFL) of 814 mm and weighed 3050 g (). The initial identification of the specimen took place on board during the scientific survey conducted by experienced researchers specializing in the species from the Porcupine Bank, including Francisco Baldó. To achieve this, they gathered data on the total length, weight, and sex of the individual, with particular emphasis on the intrinsic and distinctive characteristics of the chimaera as outlined in Ebert and Dando (Citation2020). These features comprise a very long strait conical snout, an underside of snout whitish, a mouth well in front of the eyes, a lateral head profile straight, eyes very small, a margin of upper caudal fin lobe with tubercules, a height of the upper caudal fin lobe less than height of lower caudal fin lobe and filamentous tail short. The pectoral fins are narrow, and the caudal fin exhibits an exceptionally short terminal filament and widely spaced tubercles along the upper edge in adults and large juveniles (Compagno et al. Citation1989; Ebert and Dando Citation2020). The coloration of this species ranges from whitish to light brown (Compagno et al. Citation1989). Additionally, upon reaching land, the specimen was sent to the laboratory of the Spanish Institute of Oceanography COV-CISC. There, the identification of the chimaera was confirmed by experienced personnel in Chondrichthyes, namely Nair Vilas Arrondo and Esther Roman Marcote. A total of 92 morphometric measurements were conducted following the protocols outlined by James et al. (Citation2009), Compagno (Citation1990), and Didier and Nakaya (Citation1999), as part of the framework of Nair’s doctoral thesis, which complements the taxonomic identification (Supplementary Table S1). The specimen was deposited at the Oceanographic Center of Vigo, IEO-CSIC (https://www.ieo.es/es/web/vigo/, Nair Vilas Arrondo, [email protected]) with voucher name k141_3.
Figure 1. Species reference image of Rhinochimaera atlantica. The most characteristic feature is its faction rhinoceros’ nose, in reference to the long and pointed proboscis. Photograph by Francisco Baldó onboard the R/V Vizconde de Eza during the Spanish Bottom Trawl Survey on the Porcupine Bank.

Total genomic DNA was extracted using a standard high-salt protocol (Sambrook et al. Citation1989) and sent to Macrogen (Seoul, South Korea) for Illumina Paired-End (PE) library construction (2 × 150 bp) and whole genome sequencing on a Novaseq6000 platform. Adapters, in the raw PE sequencing reads, were removed with Trimmomatic (version 0.38) by setting parameters leading and trailing to 5, minlen of 36, and a ‘sliding window’ of 4 bp with the required quality of 15 (Bolger et al. Citation2014). Mitogenome assembly and annotation were conducted with MitoZ (version 3.4) (Meng et al. Citation2019) with the clean PE reads and default parameters, using the following associated tools: GeneWise (Birney et al. Citation2004), Blast+ (Gertz et al. Citation2006), Cricos (Krzywinski et al. Citation2009), BWA (Li and Durbin Citation2009), SAMtools (Li et al. Citation2009), MiTFi (Jühling et al. Citation2012), Infernal (Nawrocki and Eddy Citation2013), HMMER (Wheeler and Eddy Citation2013), Megahit (Li et al. Citation2015), ete3 (Huerta-Cepas et al. Citation2016), Fastp (Chen et al. Citation2018), and tiara (Karlicki et al. Citation2022). The mitochondrial reads were retrieved from the whole genome sequencing outputs, by mapping the trimmed reads to the mitogenome assembly using the bbmap v.37.77 and after selecting only the mapped reads using the tool reformat.sh (implemented through bbmap) (Bushnell Citation2014). The mitogenome map was created with the annotation module of MitoZ. To generate a coverage plot, the PE reads were mapped to the final assembly using Burrows–Wheeler Aligner v.0.7.17-r1198 (Li Citation2013) and after bam2plot (https://github.com/willros/bam2plot) was used to generate the graphical plot (Supplementary Figure S2). All the available complete and partial Holocephali mitogenome assemblies (with more than 10,000 bp) were downloaded (n = 16) from GenBank (accessed on 1 December 2023). Two complete elasmobranch mitogenomes (one representing Rajiformes and one representing Squalomorphii) were also downloaded from GenBank and used as outgroup taxa.
The alignment of the 13 protein-coding genes (PCGs) retrieved from the downloaded mitogenomes was conducted with MAFFT (version 7.505) (Katoh and Standley Citation2013), trimmed with trimAL (version 1.2) (Capella-Gutiérrez et al. Citation2009) and concatenated with FasConCAT-G (version 1.05.1) (Kück and Longo Citation2014). The final alignment had a total of 11,418 bp. The identification of the partition-scheme, best-fit nucleotide substitution models, and maximum-likelihood phylogenetic inference (10,000 ultrafast bootstrap replicates) were conducted on IQ-TREE (version 1.6.12) (Nguyen et al. Citation2015; Kalyaanamoorthy et al. Citation2017).
Results
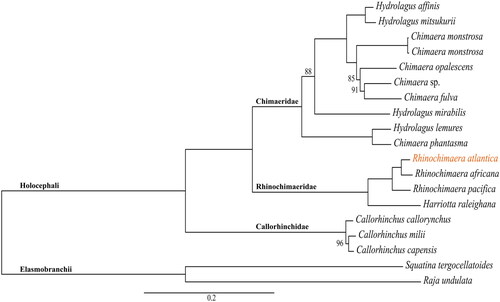
The assembled mitogenome of R. atlantica with 17,852 bp has 13 PCGs, 22 transfer RNA, and two ribosomal RNA genes (). Nine of these genes (one PCG, NADH dehydrogenase subunit 6 (ND6), and eight transfer RNA – trnP(ugg), trnQ(uug), trnA(ugc), trnN(guu), trnC(gca), trnY(gua), trnS(uga), and trnE(uuc)) are in the complementary strand. All PCGs started with the ATG codon, except for the cytochrome c oxidase subunit 1 (COX1), which started with GTG. The nucleotide composition of the R. atlantica mitogenome is A = 30%, T = 28.5%, G = 14.3%, and C = 27.2%, with a GC content of 41.5%. Overlap of sequences was detected in eight mitochondrial genes, ranging between 1 and 13 bp. The mitogenome has been deposited in GenBank under accession number OQ947382. The phylogenetic inference () demonstrates the three Holocephali families to be monophyletic, and that Chimaeridae and Rhinochimaeridae are sister groups with strong support (100% bootstrap probabilities). R. atlantica is placed in the Rhinochimaeridae clade.
Figure 2. Mitogenome map of Rhinochimaera atlantica with 17,852 bp, 13 protein-coding genes (ND1, ND2, COX1, COX2, ATP8, ATP6, COX3, ND3, ND4L, ND4, ND5, ND6, and CYTB), 22 transfer RNA (beginning by ‘trn’), and two ribosomal RNA genes (s-rRNA and l-rRNA). Nine of these genes (one protein-coding gene (ND6) and eight transfer RNA – trnP(ugg), trnQ(uug), trnA(ugc), trnN(guu), trnC(gca), trnY(gua), trnS(uga), and trnE(uuc)) are in the complementary strand, demonstrated in the outside of the circle. The inner circle represents the coverage plot (blue).
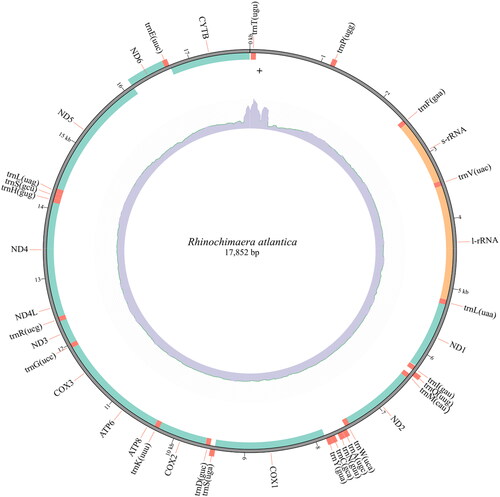
Figure 3. Maximum-likelihood phylogenetic inference with the sequences of all protein-coding genes from all available Holocephali mitogenomes (n = 17). The sequences were downloaded from GenBank (accessed on 1 December 2023) with the following accession numbers: MT090368 (H. affinis) (Gomes-dos-Santos et al. Citation2020), OP391486 (H. mitsukurii) (Wang et al. Citation2023), MT410927 (C. monstrosa) (unpublished), AJ310140 (C. monstrosa) (Arnason et al. Citation2001), OK638184 (C. opalescens) (Vilas-Arrondo et al. Citation2022), MT880605 (Chimaera sp.) (unpublished), HM147138 (C. fulva) (Inoue et al. Citation2010), MW029477 (H. mirabilis) (Gomes-dos-Santos et al. Citation2021), HM147139 (H. lemures) (Inoue et al. Citation2010), KP006329 (C. phantasma) (unpublished), KP006330 (R. africana) (unpublished), HM147141 (R. pacifica) (Inoue et al. Citation2010), HM147140 (H. raleighana) (Inoue et al. Citation2010), HM147135 (C. callorhynchus) (Inoue et al. Citation2010), HM147137 (C. milii) (Inoue et al. Citation2010), and HM147136 (C. capensis) (Inoue et al. Citation2010). Outgroup taxa: ON210850 (S. tergocellatoides) (unpublished) and MT274574 (R. undulata) (Kousteni et al. Citation2021). The mitogenome of R. atlantica provided in this study was also added to this phylogenetic inference (accession number: OQ947382). Only the bootstrap support values lower than 100 are represented in the figure.
Discussion and conclusions
The length of the R. atlantica mitogenome is within the range of the two available Rhinochimaera mitogenomes (one from R. pacifica (24,889 bp) and one from R. africana (15,446 bp)) (Inoue et al. Citation2010). A long noncoding region (between trnT(ugu) and trnP(ugg) genes) with variable size has been described in holocephalans mitogenomes and pointed as the main contributor to the size variation observed in these mitogenomes (Inoue et al. Citation2010). In the R. atlantica mitogenome sequence provided here, this region is present with a size of 1086 bp. Moreover, this well-reported mitochondrial feature offers challenges to the genome assembler thus explaining the positive coverage variation observed (Supplementary Figure S2). In vertebrates, the control region (CR) is located within the tRNA-Phe and tRNA-Pro (Satoh et al. Citation2016). In our assembly, this region is well resolved, it has 1222 bp () and has a similar size to other mitogenome assemblies from the same family Rhinochimaeridae (e.g. 1304 bp in Rhinochimaera pacifica – HM147141.1; 1316 bp in Harriotta raleighana – HM147140.1).
In the phylogeny provided here (), R. atlantica is in the Rhinochimaeridae clade, as expected. This clade has a group, recovered with 100% bootstraps probabilities, with all Rhinochimaera species (including our R. atlantica mitogenome), and Harriotta raleighana as its sister species (). Rhinochimaeridae encompasses three genera, Rhinochimaera, Harriotta, and Neoharriotta, with the latter suggested to be the most basal of these genera (Licht et al. Citation2012). However, in our phylogenetic analysis, only the mitogenomes of Rhinochimaera and Harriotta were considered as no Neoharriotta mitogenomes are presently available. Comparatively, Chimaeridae in this phylogeny has a wider taxonomic representation ().
We provide the first complete R. atlantica mitogenome. Among Chondrichthyes, there is a lack of molecular data for Holocephali. This mitogenome will contribute to the genomic characterization of R. atlantica. The need to produce more complete Holocephali mitogenomes is highlighted.
Author contributions
LC and EF conceived and designed the research. FB and JMA performed sample collection and specimen validation. NVA conducted DNA extraction. AM, NVA, AGS, AV, ERM, FB, JMA, MP, and MLL performed and discussed phylogenetic analysis. AM, AGS, and MLL conducted the mitogenome assembly. All authors interpreted and discussed the data. All authors were involved in the draft preparation. All authors read, edited, and approved the final manuscript. All authors agree to be accountable for all aspects of the work. AM is the author who handled the correspondence with the editorial office.
Ethical approval
The described work was approved by the CIIMAR Ethical Committee and CIIMAR Managing Animal Welfare Body (ORBEA), according to the European Union Directive 2010/63/EU. R. atlantica voucher specimen has been deposited at the Oceanographic Center of Vigo, IEO-CSIC (contact person: Nair Vilas Arrondo, [email protected]) with voucher name k141_3.
Supplemental Material
Download MS Word (612.1 KB)Supplemental Material
Download PNG Image (592.1 KB)Supplemental Material
Download PNG Image (4.2 MB)Acknowledgements
We would like to thank the staff involved in the research survey PORCUPINE 2021 of the Spanish Institute of Oceanography (IEO, CSIC) on board the R/V Vizconde de Eza (Ministry of Agriculture, Fisheries and Food, Spain).
Disclosure statement
The author Manuel Lopes-Lima is an Editorial Board Member for the journal – TMDN.
Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI at https://www.ncbi.nlm.nih.gov under the accession number OQ947382. The associated BioProject, SRA, and Bio-Sample numbers are PRJNA1055343, SRR27310001, and SAMN38990799, respectively.
Additional information
Funding
References
- Arnason U, Gullberg A, Janke A. 2001. Molecular phylogenetics of gnathostomous (jawed) fishes: old bones, new cartilage. Zool Scripta. 30(4):249–255. doi:10.1046/j.1463-6409.2001.00067.x.
- Barnett LAK, Ebert DA, Cailliet GM. 2012. Evidence of stability in a chondrichthyan population: case study of the spotted ratfish Hydrolagus colliei (Chondrichthyes: Chimaeridae). J Fish Biol. 80(5):1765–1788. doi:10.1111/j.1095-8649.2011.03216.x.
- Birney E, Clamp M, Durbin R. 2004. GeneWise and Genomewise. Genome Res. 14(5):988–995. doi:10.1101/gr.1865504.
- Bolger AM, Lohse M, Usadel B. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 30(15):2114–2120. doi:10.1093/bioinformatics/btu170.
- Bushnell B. 2014. BBMap: a fast, accurate, splice-aware aligner. LBNL Report LBNL-7065E, Lawrence Berkeley National Laboratory. Berkeley, CA.
- Capella-Gutiérrez S, Silla-Martínez JM, Gabaldón T. 2009. trimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics. 25(15):1972–1973. doi:10.1093/bioinformatics/btp348.
- Carpenter JS. 1969. First record of the long-snouted ratfish, Rhinochimaera atlantica in the Gulf of Mexico. Copeia. 1969(1):203–204. doi:10.2307/1441723.
- Carpenter KE. 2002. The living marine resources of the Western Central Atlantic. Volume 1: introduction, molluscs, crustaceans, hagfishes, sharks, batoid fishes, and chimaeras. FAO Species Identification Guide for Fishery Purposes and American Society of Ichthyologists and Herpetologists Special Publication No. 5; p. 1–600.
- Chen S, Zhou Y, Chen Y, Gu J. 2018. fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics. 34(17):i884–i890. doi:10.1093/bioinformatics/bty560.
- Compagno LJ. 1990. Alternative life-history styles of cartilaginous fishes in time and space. Environ Biol Fish. 28(1–4):33–75. doi:10.1007/BF00751027.
- Compagno LJV, Didier DA, Burgess GH. 2005. Classification of Chondrichthyan fish. In: Fowler SL, Cavanagh RD, Camhi M, editors. Sharks, rays and chimaeras: the status of the chondrichthyan fishes. Gland, Switzerland; Cambridge (UK): IUCN; p. 4–11.
- Compagno LJV, Ebert DA, Smale MJ. 1989. Guide to the sharks and rays of Southern Africa. London: New Holland (Publ.) Ltd.; p. 158.
- Didier DA, Kemper JM, Ebert DA. 2012. Phylogeny, biology, and classification of extant holocephalans. In: Carrier JC, Musick JA, Heithaus MR, editors. Marine biology. 2nd ed. Boca Raton: CRC Press, Taylor & Francis Group; p. 97–122.
- Didier DA, Nakaya K. 1999. Redescription of Rhinochimaera pacifica (Mitsukuri) and first record of R. africana Compagno, Stehmann & Ebert from Japan (Chimaeriformes: Rhinochimaeridae). Ichthyol Res. 46(2):139–152. doi:10.1007/BF02675432.
- Ebert DA, Dando M. 2020. Field guide to sharks, rays & chimaeras of Europe and the Mediterranean. Vol. 20. Princeton: Princeton University Press.
- Finucci B, Dunn MR, Jones EG, Anderson J. 2017. Reproductive biology of the two deep-sea chimaerids, longnose spookfish (Harriotta raleighana) and Pacific spookfish (Rhinochimaera pacifica). Deep Sea Res Part I. 120:76–87. doi:10.1016/j.dsr.2016.11.008.
- Finucci B. 2020. Rhinochimaera atlantica. The IUCN red list of threatened species. e.T60145A124444407. doi:10.2305/IUCN.UK.2020-2.RLTS.T60145A124444407.en.
- Gertz EM, Yu YK, Agarwala R, Schäffer AA, Altschul SF. 2006. Composition-based statistics and translated nucleotide searches: improving the TBLASTN module of BLAST. BMC Biol. 4:1–14. doi:10.1186/1741-7007-4-41.
- Gomes-dos-Santos A, Arrondo NV, Machado AM, Veríssimo A, Pérez M, Román E, Castro LFC, Froufe E. 2020. The complete mitochondrial genome of the deep-water cartilaginous fish Hydrolagus affinis (de Brito Capello, 1868) (Holocephali: Chimaeridae). Mitochondrial DNA Part B. 5(2):1810–1812. doi:10.1080/23802359.2020.1749154.
- Gomes-dos-Santos A, Vilas-Arrondo N, Machado AM, Veríssimo A, Pérez M, Baldó F, Castro LFC, Froufe E. 2021. Shedding light on the Chimaeridae taxonomy: the complete mitochondrial genome of the cartilaginous fish Hydrolagus mirabilis (Collett, 1904) (Holocephali: Chimaeridae). Mitochondrial DNA B Resour. 6(2):420–422. doi:10.1080/23802359.2020.1870887.
- Huerta-Cepas J, Serra F, Bork P. 2016. ETE 3: reconstruction, analysis, and visualization of phylogenomic data. Mol Biol Evol. 33(6):1635–1638. doi:10.1093/molbev/msw046.
- Iglésias SP, Kemper JM, Naylor GJ. 2022. Chimaera compacta, a new species from southern Indian Ocean, and an estimate of phylogenetic relationships within the genus Chimaera (Chondrichthyes: Chimaeridae). Ichthyol Res. 69(1):31–45. doi:10.1007/s10228-021-00810-9.
- Inoue JG, Miya M, Lam K, Tay B-H, Danks JA, Bell J, Walker TI, Venkatesh B. 2010. Evolutionary origin and phylogeny of the modern holocephalans (Chondrichthyes: Chimaeriformes): a mitogenomic perspective. Mol Biol Evol. 27(11):2576–2586. doi:10.1093/molbev/msq147.
- James KC, Ebert DA, Long DJ, Didier DA. 2009. A new species of chimaera, Hydrolagus melanophasma sp. nov. (Chondrichthyes: Chimaeriformes: Chimaeridae), from the Eastern North Pacific. Zootaxa. 2218(1):59–68. doi:10.11646/zootaxa.2218.1.3.
- Jühling F, Pütz J, Bernt M, Donath A, Middendorf M, Florentz C, Stadler PF. 2012. Improved systematic tRNA gene annotation allows new insights into the evolution of mitochondrial tRNA structures and into the mechanisms of mitochondrial genome rearrangements. Nucleic Acids Res. 40(7):2833–2845. doi:10.1093/nar/gkr1131.
- Kalyaanamoorthy S, Minh BQ, Wong TKF, Von Haeseler A, Jermiin LS. 2017. ModelFinder: fast model selection for accurate phylogenetic estimates. Nat Methods. 14(6):587–589. doi:10.1038/nmeth.4285.
- Karlicki M, Antonowicz S, Karnkowska A. 2022. Tiara: deep learning-based classification system for eukaryotic sequences. Bioinformatics. 38(2):344–350. doi:10.1093/bioinformatics/btab672.
- Katoh K, Standley DM. 2013. MAFFT Multiple Sequence Alignment Software Version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780. doi:10.1093/molbev/mst010.
- Kousteni V, Mazzoleni S, Vasileiadou K, Rovatsos M. 2021. Complete mitochondrial DNA genome of nine species of sharks and rays and their phylogenetic placement among modern elasmobranchs. Genes. 12(3):324. doi:10.3390/genes12030324.
- Krzywinski M, Schein J, Birol I, Connors J, Gascoyne R, Horsman D, Jones SJ, Marra MA. 2009. Circos: an information aesthetic for comparative genomics. Genome Res. 19(9):1639–1645. doi:10.1101/gr.092759.109.
- Kück P, Longo GC. 2014. FASconCAT-G: extensive functions for multiple sequence alignment preparations concerning phylogenetic studies. Front Zool. 11(1):81. doi:10.1186/s12983-014-0081-x.
- Li D, Liu CM, Luo R, Sadakane K, Lam TW. 2015. MEGAHIT: an ultra-fast single-node solution for large and complex metagenomics assembly via succinct de Bruijn graph. Bioinformatics. 31(10):1674–1676. doi:10.1093/bioinformatics/btv033.
- Li H, Durbin R. 2009. Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics. 25(14):1754–1760. doi:10.1093/bioinformatics/btp324.
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R, 1000 Genome Project Data Processing Subgroup. 2009. The sequence alignment/map format and SAMtools. Bioinformatics. 25(16):2078–2079. doi:10.1093/bioinformatics/btp352.
- Li H. 2013. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv preprint arXiv:1303.3997v2.
- Licht M, Schmuecker K, Huelsken T, Hanel R, Bartsch P, Paeckert M. 2012. Contribution to the molecular phylogenetic analysis of extant holocephalan fishes (Holocephali, Chimaeriformes). Org Divers Evol. 12(4):421–432. doi:10.1007/s13127-011-0071-1.
- Meng G, Li Y, Yang C, Liu S. 2019. MitoZ: a toolkit for animal mitochondrial genome assembly, annotation and visualization. Nucleic Acids Res. 47(11):e63. doi:10.1093/nar/gkz173.
- Nawrocki EP, Eddy SR. 2013. Infernal 1.1: 100-fold faster RNA homology searches. Bioinformatics. 29(22):2933–2935. doi:10.1093/bioinformatics/btt509.
- Nguyen LT, Schmidt HA, Von Haeseler A, Minh BQ. 2015. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 32(1):268–274. doi:10.1093/molbev/msu300.
- Sambrook J, Fritsch ER, Maniatis T. 1989. Molecular cloning: a laboratory manual. New York: Cold Harbor Spring Press.
- Satoh TP, Miya M, Mabuchi K, Nishida M. 2016. Structure and variation of the mitochondrial genome of fishes. BMC Genomics. 17(1):719. doi:10.1186/s12864-016-3054-y.
- Stevens JD, Bonfil R, Dulvy NK, Walker PA. 2000. The effects of fishing on sharks, rays, and chimaeras (chondrichthyans), and the implications for marine ecosystems. ICES J Mar Sci. 57(3):476–494. doi:10.1006/jmsc.2000.0724.
- Vilas-Arrondo N, Gomes-Dos-Santos A, Pérez M, Baldó F, Veríssimo A, Catarino D, Machado AM, Román-Marcote E, Bañón R, Froufe E, et al. 2022. A mitochondrial genome assembly of the opal chimaera, Chimaera opalescens Luchetti, Iglésias et Sellos 2011, using PacBio HiFi long reads. Mitochondrial DNA B Resour. 7(3):434–437. doi:10.1080/23802359.2022.2044403.
- Wang X, Fang C, Wang C. 2023. Complete mitochondrial genome of Hydrolagus mitsukurii (Jordan & Snyder, 1904). Mitochondrial DNA B Resour. 8(6):682–685. doi:10.1080/23802359.2023.2222852.
- Weigmann S. 2016. Annotated checklist of the living sharks, batoids and chimaeras (Chondrichthyes) of the world, with a focus on biogeographical diversity. J Fish Biol. 88(3):837–1037. doi:10.1111/jfb.12874.
- Wheeler TJ, Eddy SR. 2013. nhmmer: DNA homology search with profile HMMs. Bioinformatics. 29(19):2487–2489. doi:10.1093/bioinformatics/btt403.
- Yamaguchi K, Uno Y, Kadota M, Nishimura O, Nozu R, Murakumo K, Matsumoto R, Sato K, Kuraku S. 2023. Elasmobranch genome sequencing reveals evolutionary trends of vertebrate karyotypic organization. Genome Res. 33(9):1527–1540. doi:10.1101/gr.276840.122.
