Abstract
The majority of the Origanum species are important medicinal plants as well as culinary herbs and have thus a great economic value. Some taxonomic issues are still pending within the genus and the cytogenetic studies about this genus are still very scarce. Therefore, studies concerning chromosome number and genome size can provide complementary data that may be useful to characterize the genus Origanum. These two approaches have been used to characterize five Moroccan taxa of the genus Origanum occurring in the wild in addition to the exotic species O. onites. All investigated taxa are diploid with chromosome number of 2n = 30. This is the first time the chromosome numbers have been counted in O. grosii, O. compactum and O. × font-queri as well as in O. vulgare subsp. virens from Morocco. The genome sizes are considered as small, and the mean values ranged from 1.43 pg/2C in O. vulgare subsp. virens to 1.53 pg/2C in O. compactum. Besides, no significant intraspecific variability in genome size was observed among populations of O. elongatum as well as of O. grosii.
Introduction
Morocco is one of the most interesting floristic areas in North Africa. The flora of Morocco is estimated to comprise 978 endemic taxa, which constitute more than half of North African endemic plants (El Oualidi et al. Citation2012). This endemic richness may be due to the presence of mixed and well differentiated environments. Origanum is one of the genera that contain important endemic plant taxa in the country.
The genus Origanum L. (Lamiaceae), known as “zaatar” in Morocco, is characterized by a remarkable morphological and chemical diversity (Aboukhalid et al. Citation2016; Bakhy et al. Citation2014). This biodiversity is reflected by the existence of 67 taxa including 18 hybrids, (Carlström Citation1984; Danin Citation1990; Danin and Künne Citation1996; Duman, Baser, and Aytec Citation1998; Duman et al. Citation1995; Ietswaart Citation1980), most of which have a circum-Mediterranean distribution (Kokkini Citation1997).
The center of diversity of the genus Origanum is considered to be in the Eastern part of the Mediterranean with 21 taxa occur in Turkey (Kokkini Citation1997). The flora of Morocco comprises six taxa of Origanum (belonging to four sections) three of which are endemic: O. grosii Pau & Font Quer ex Ietsw., O. elongatum (Bonnet) Emb. & Maire and O. × font-queri Pau (hybrid O. compactum × O. grosii). Moreover, O. compactum Benth. is endemic to Morocco and South Spain, O. vulgare subsp. virens (Hoffmanns. & Link) Ietsw. is indigenous to the Western Mediterranean, and O. majorana L. is present in Morocco as a cultivated species (El Oualidi and Navarro Citation2007; Valdés et al. Citation2002). Due to their very similar morphologies, the three Moroccan steno-endemics O. elongatum, O. grosii (section Elongatispica Ietsw.) and O. × font-queri are subject to taxonomic confusion. According to the Euro+Med (Citation2006) Plant database and African Plant Database (version 3.4.0) (Dobignard and Chatelain Citation2010‒13), O. grosii and O. × font-queri are considered as synonyms of O. elongatum. However, Ietswaart (Citation1980) described O. grosii and O. elongatum as two different Origanum species according to their morphological characters with the stem, spike and leaf lengths of O. elongatum being larger than those of O. grosii, but with O. grosii having a longer and broader bract than O. elongatum, while the hybrid, O. × font-queri, is not described in the classification of Ietswaart (Citation1980). On the other hand, both O. elongatum and O. grosii present obvious morphological differences with respect to O. compactum (section Prolaticorolla Ietsw.) as well as to O. vulgare subsp. virens (section Origanum L.).
The geographical distribution range of indigenous oregano in Morocco varies from species to species. O. compactum is the most common, followed by O. elongatum with a very large area of distribution, and O. grosii and O. vulgare subsp. virens with a more restricted area. Due to their economic importance related to therapeutic and culinary characteristics, all populations of oregano taxa in Morocco are under intensive exploitation, a situation that has triggered a reduction of their abundance and caused complete disappearance in some areas in Morocco, especially for O. compactum and O. vulgare subsp. virens.
Cytogenetics plays an important role in the evaluation of inter and intra-specific variability, and in the detection of the structural changes in the genome (Mas de Xaxars et al. Citation2015; Yang et al. Citation2009). Besides, flow cytometry is a valuable approach that permits the rapid and precise screening of nuclear DNA genome size and ploidy level and can complement data coming from classical cytogenetics (Siljak-Yakovlev et al. Citation2008).
Contrary to the important investigations in the field of chemistry and molecular (Azizi, Yan, and Honermeier Citation2009; Lukas, Schmiderer, and Novak Citation2013; Skoula et al. Citation1999), the cytogenetic studies about Origanum are restricted and have been mainly focused on chromosome number determination (Balim and Kesercioğlu Citation1998; Lepper Citation1970). Cytogenetic data concerning the karyotype, ploidy level or genome size of Origanum species are especially very scarce for the species from Morocco.
The purpose of this study was (1) to characterize the genome of all six Origanum taxa growing in Morocco by chromosome number counting, ploidy level and genome size assessment, and (2) to detect any genome size variability among populations of the two endemic species O. elongatum and O. grosii, according to their geographical distribution.
Material and methods
Plant material
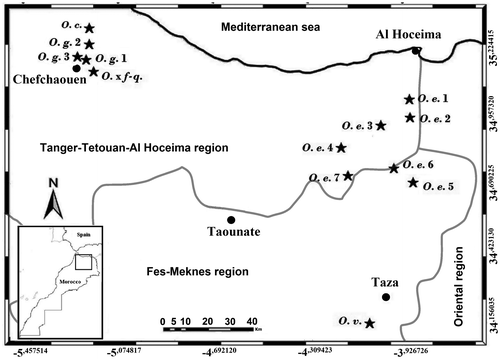
The plant material of the five Origanum taxa investigated in this study was collected among wild populations from Morocco in August 2015. In addition, the exotic O. onites was also studied (Figure ). Taxonomic identification and verification were performed following the key of Origanum taxonomy of Ietswaart (Citation1980). Besides, the hybrid O. × font-queri collected from areas displaying neighboring populations of O. compactum and O. grosii., was determined according to its intermediate characters between its parent taxa. The geographical data of each studied population is presented in Table , and Figure shows the locations of the populations on the map. Voucher specimens were deposited at the herbarium of the Scientific Institute of Rabat (SIR), Morocco.
Figure 1. Stems with flowering of six studied Origanum taxa transplanted in the experimental station of INRA-Morocco (A) O. compactum; (B) O. onites; (C) O. elongatum; (D) O. vulgare subsp. virens; (E) O. grosii; (F) O. × font-queri.
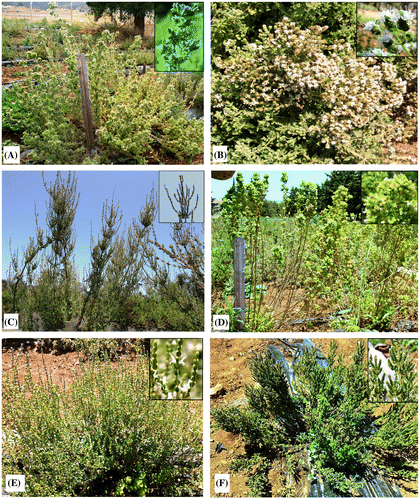
Table 1. Geographical description of studied populations.
Germination of seeds and karyological technique
The seeds isolated from mature inflorescences were first disinfected with sodium hypochlorite (10%), washed in distilled water and placed to germinate in Petri dishes at 22 °C within a two-week period. The germinated seedling with well-developed root tips of about 1 cm length were pre-treated with 0.05% colchicine (Sigma Chemical Co.) solution for 60 min at 22 °C or with 0.002 M 8-hydroxyquinoline (Merck or Prolabo) for 3 h at 16 °C. Fixation was performed in 3:1 (v/v) absolute ethanol: acetic acid for at least 24–48 h at 4 °C. The material was then stored in 70% ethanol at 4 °C. Subsequently, the meristems of root tips were hydrolysed in 1 N HCl for 6 min at 60° C, and stained in Schiff reagent following the standard Feulgen and Rossenbeck (Citation1924) method. The squash was performed in a drop of acetic carmine or acetic orcein.
Flow cytometry
The total nuclear DNA amount was assessed by flow cytometry according to Marie and Brown (Citation1993). Young leaves of the analysed individuals and the internal standard (Solanum lycopersicum L. (Montfavet 63–5 (2C = 1.99 pg, Lepers-Andrzejewski et al. Citation2011)) were chopped together using a razor blade in a plastic Petri dish with 1 ml of Gif nuclei-isolation buffer (45 mM MgCl2, 30 mM sodium citrate, 60 mM MOPS (4-morpholine propane sulfonate pH 7), 1% (w/v) polyvinylpyrrolidone 10,000, pH 7.2) containing 0.1% (w/v) Triton X–100, supplemented with 5 mM sodium metabisulfite and RNAse (2.5 U/ml). The suspension was filtered through a 50 μm nylon mesh. The nuclei were stained with 50 μg/ml propidium iodide, a specific DNA intercalating fluorochrome dye, and kept at least 5 min at 4 °C. The 5000–10,000 stained nuclei were measured for each sample using a flow cytometer with a 532 nm 30 mW laser (CyFlow SL3, Partec, Munster, Germany). The total holoploid nuclear DNA content (2C) was calculated using the linear relationship between the fluorescent signals from the stained nuclei of the Origanum species and those of the internal standard. The mean 2C-value, as well as the standard deviation of the mean and its coefficient of variation (%), were calculated from measurements of samples comprising 2 to 5 individuals depending on the population.
Statistical analysis
The inter-specific genome size differences among the six studied taxa and the inter-populational variability among all populations were analysed using ANOVA with the Welch test for unequal sample (Welch Citation1947). On the other hand, the intra-specific variation for two of the species (O. elongatum and O. grosii) were tested by both ANOVA test for equal sampled sizes (O. elongatum) and ANOVA with the Welch test for unequal samples (O. grosii). A multiple comparison test (Games-Howell test) has been applied to indicate pair-wise differences in the six studied taxa. All tests were performed by the Statistical Analysis System Software (SAS) version 9.2.
Results
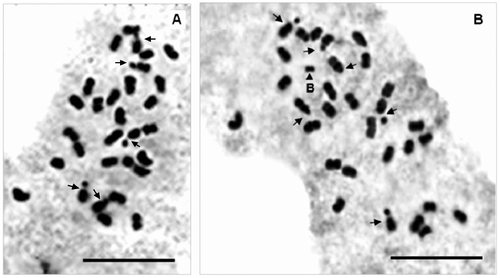
The results concerning somatic chromosome numbers (2n), ploidy level (x) and genome size (2C) of each studied population are presented in Table . The chromosome number determined for all here studied Origanum taxa and populations was 2n = 30 (Figure ), with the basic chromosome number x = 15. Moreover, one B chromosome and 6 satellite chromosomes were observed in mitotic metaphase of population 7 of O. elongatum (Figure ).
Table 2. Summary of results concerning genome size assessment.
Figure 3. Mitotic metaphase plates of Origanum elongatum (O. e. 7) 2n = 30 (A) and 2n = 30+B (B). B chromosome is indicated with arrowhead and satellite chromosome with arrows. Bar scale = 10μm.
In view of the genome size, the mean nuclear 2C DNA content of all species was 1.48 pg. The 2C value varied from 1.43 pg in O. vulgare subsp. virens to 1.53 pg in O. compactum. Regarding the intra-specific variation in O. elongatum and O. grosii, the difference of genome size among populations was quite small (0.02 pg) and ranged from 1.48 to 1.50 pg in O. elongatum, and from 1.46 to 1.48 pg in O. grosii. ANOVA with the Welch test for 2C DNA content (pg) revealed significant inter-specific and inter-populational differences (F = 23,665, p = 0.000 and F = 9,445 p = 0.000 respectively). The pair-wise comparisons in the six studied taxa by Games-Howell analysis (see Table ) showed that there was no significant difference between cultivated O. onites and all the other taxa, whereas O. vulgare subsp. virens showed significant difference with the other taxa. In addition, there was a high significant difference between O. × font-queri and O. elongatum as well as between O. × font-queri and O. grosii (Table , p = 0.000).
Table 3. Multiple comparisons (dependent variable: genome size (pg)), with the average difference between the studied taxa and standard error.
The studied populations of O. elongatum occur along eastern part of Rif Mountains from 681 to 1815 m and climate varies from semi-arid to perhumid. Concerning the intra-specific variation, the genome size ranged from 1.45 pg to 1.52 pg with a mean value of 2C = 1.49 pg. In contrast, the populations of O. grosii occurring in the western part of Rif (402 – 1070 m) and in habitats characterized by humid climate, showed almost the same range of genome size variation from 1.45 to 1.5 pg, with a mean value of 2C = 1.47 pg.
No significant genome size difference was identified between O. elongatum and O. grosii. While the hybrid O. × font-queri (O. compactum × O. grosii) showed an intermediate 2C value between the parent taxa, 1.52 pg (mean), closer to O. compactum (1.53 pg) than to O. grosii (1.47 pg).
Table summarizes the principal variability of morphological characteristics of the Moroccan Origanum species according to Ietswaart (Citation1980).
Table 4. Comparative morphological characteristics of four Moroccan Origanum taxa according to Ietswaart (1980).
Discussion
Chromosome number
The results indicate that all taxa are diploid and that there is no difference in chromosome number within the studied panel of Origanum species. According to previous studies cited on the Chromosome Counts Data Base (CCDB) (Rice et al. Citation2014) and Index to Plant Chromosome Numbers (IPCN) (Goldblatt and Johnson Citation1979) databases, Origanum taxa investigated until now also presented the same chromosome number 2n = 30, even though they belong to different sections and are growing in different regions. The only divergence in chromosome number is recorded in O. vulgare which is characterized by three different chromosomes numbers 2n = 28, 2n = 30 and 2n = 32 (Table ). This variability seems to be due to the aneuploidy.
Table 5. The chromosome number of all investigated Origanum taxa according to the databases CCDB (Chromosome counts Data Base) and IPCN (Index to Plant Chromosome Numbers).
The chromosome number 2n = 30 was reported frequently in the Origanum related genera, like Micromeria sp. (Morales Citation1990), Thymus sp. (Mehrpur et al. Citation2002) and Satureja sp. (Boscaiu et al. Citation2000). Nevertheless, many studies have also reported heterogeneous chromosome numbers such as in Micromeria inodora (Desf.) Benth.: 2n = 26 (Cardona Citation1973), Thymus maroccanus Ball.: 2n = 24 (Tahiri, Rejdali, and Atbib Citation1998), and Satureja hortensis L.: 2n = 24 (Gill Citation1981). Moreover, some species of Thymus and Micromeria displayed polyploidy in some areas of their geographical distribution [e.g. Thymus kotschyanus Boiss. & Hohen.: 2n = 4x = 60 (Mehrpur et al. Citation2002) and Micromeria graeca (L.) Benth.: 2n = 4x = 60 (Luque and Lifante Citation1991)], which was not the case for Origanum investigated taxa until present.
Inter- and intra-specific genome size variation
The genome size has been estimated for the first time in all the studied taxa according to Plant DNA C-values database (release 6.0, Dec. 2012) (Bennett and Leitch Citation2012). According to Leitch et al. (Citation2005), the estimated genome sizes of the studied taxa are considered as very small (2C ≤ 2.8 pg). Only three species (out of 49) of genus Origanum have been measured until now. O. vulgare L. with 2C = 1.36 pg (Mowforth Citation1986) and O. libanoticum Boiss. with 2C = 1.44 pg (Bou Dagher-Kharrat et al. Citation2013) present smaller genome size in comparison with those of the present study. In contrary, O. syriacum L. had a bigger genome size (2C = 1.64 pg, Bou Dagher-Kharrat et al. Citation2013).
Moreover, these present values indicate that there is little variation in genome size among the populations of the same species and among individuals of the same population (Table ), since no significant difference was identified among populations of O. elongatum as well as of O. grosii (p > 0.05). It appears that neither the climate nor the altitude influenced the variation in genome size of the present populations of the studied species. The results show also that all studied populations are diploid and no polyploidization pattern was observed according geographical distribution. Within the investigated individuals (from all populations), O. vulgare subsp. virens had the bigger differences between maximal and minimal 2C DNA values (0.10 pg), followed by O. onites (0.08 pg), O. compactum, O. elongatum (0.07 pg), and O. grosii (0.05 pg) while the hybrid O. × font-queri had the smaller one (0.01 pg).
The hybrid O. × font-queri appears as a result of gene flow between its parent taxa, O. grosii and O. compactum. This hybrid was determined for the first time by Pau in 1930, but no botanical description was given. The hybrid is close to O. grosii in the morphological characters, and characterized by larger bracts and calyx than in O. grosii as well as by its more or less glabrous stems and leaves. The geographical and the morphological data support the possibility of occurrence of O. × font-queri at the junction of populations from parent species. However, the hybrid has a genome size (2C value) between its parent taxa, with significant variability compared with O. grosii.
The comparison of the genome size of Origanum with those of the related genera, according to Plant DNA C-values database (release 6.0, Dec. 2012) (Bennett and Leitch Citation2012), showed that all the investigated species of the genus Satureja have a bigger genome size than Origanum with a mean 2C value of 3.9 pg and that all the investigated species of Micromeria have a smaller genome size than Origanum with a mean 2C value of 0.86 pg, while the genus Thymus is considered as the closest to Origanum with a mean 2C value of 1.68 pg.
The results obtained in this study contribute to characterize six Origanum taxa at the cytogenetic level. This is the first time the chromosomes numbers are counted in O. grosii, O. compactum and O. × font-queri. In addition, the chromosome set in O. vulgare subsp. virens is counted for the first time in Morocco. Despite their high morphological and chemical variability as well as the large geographic range of distribution, all investigated Origanum species have the same chromosome number and ploidy level, and more or less similar genome size. There was no consistent pattern of significant genome size variation among populations regardless of geographic origin and altitudinal profile. In fact, the genus Origanum is considered one of the most conserved genera within the Lamiaceae family at the genetic scale. The variability of the chloroplast DNA within the genus Origanum is extremely low (Lukas and Novak Citation2013). Besides the phylogenetic studies showed low divergence between the different Origanum studies taxa (Bräuchler, Meimberg, and Heubl Citation2010; CitationLukas et al. 2013). The genus has undergone considerable rate of hybridization between several species where 18 hybrids appeared (Ietswaart Citation1980). In this context, the cytogenetic data provides important complementary parameters in the revision of many plants groups within the Lamiaceae family such as Sideritis (Esra, Duman, and Ünal Citation2008). Our study has increased significantly the data on chromosome numbers (now 17 among 67) and genome size of Origanum taxa (9); however, more investigations are needed to expand the knowledge and be able to draw solid conclusions.
Notes on contributors
Mohamed Bakha Ph.D. student, Laboratory of Biology and Health, Faculty of Sciences of Tétouan, Abdelmalek Essaâdi University, Morocco. Research Unit for Aromatic and Medicinal Plants, I.N.R.A.-Rabat, Morocco. Contribution: collected the plant materials, conceived and designed the study, performed the research experiments, wrote the manuscript and approved the final version of manuscript.
Chaouki Al Faiz Ph.D., I.N.R.A. researcher, President of the Department of Improvement and Conservation of Genetic Resources I.N.R.A., Morocco. Conservation, improvement and domestication of aromatic and medicinal plants. Contribution: participated in the collection of the plant materials, conceived and designed the study, read and approved the final version of manuscript.
Marwa Daoud Ph.D. student, Center for Ecology and Conservation Sciences (C.E.S.C.O.), National Museum of Natural History, Paris, France. Contribution: conceived and designed the study, performed the research experiments, wrote the manuscript and approved the final version of manuscript.
Noureddine El Mtili Ph.D., Professor, Laboratory of Biology and Health, Faculty of Sciences of Tétouan, Abdelmalek Essaâdi University, Morocco. Contribution: conceived and designed the study, read and approved the final version of manuscript.
Kaoutar Aboukhalid Ph.D. student, Laboratory of Applied Chemistry and Environment, Faculty of Sciences and Techniques, Hassan I University, Settat, Morocco. Research Unit for Aromatic and Medicinal Plants, I.N.R.A.-Rabat, Morocco. Contribution: participated in the collection of the plant materials, read and approved the final version of manuscript.
Abdelkarim Khiraoui Ph.D. student, Team of Environment and Valorization of Agro-Resources, Faculty of Sciences and Technologies, University, Beni Mellal, Morocco. Research Unit for Aromatic and Medicinal Plants, I.N.R.A.-Rabat, Morocco. Contribution: participated in the collection of the plant materials, read and approved the final version of manuscript.
Nathalie Machon Professor and director of research (C.E.S.C.O.), Center for Ecology and Conservation Sciences, National Museum of Natural History, Paris, France. Main research: Conservation of rare plants, Urban ecology. Contribution: funded, conceived and designed the study, read and approved the final version of manuscript.
Sonja Siljak-Yakovlev Ph.D., C.N.R.S. researcher since 1977, director of research since 1992, at present director of research emeritus, university Paris-Saclay, Ecology, Systematics, Evolution U.M.R. 8079 U.P.S.-C.N.R.S.-AgroParisTech, Department “Evolution of Angiosperms”. Main research field: Chromosome evolution and genome organization, Evolution of plants, Biodiversity, Endemism, Plant Evolutional Systematic. Contribution: funded, conceived and designed the study, performed the research experiments, wrote the manuscript and approved the final version of manuscript.
Disclosure statement
No potential conflict of interest was reported by the authors.
Acknowledgements
We would like to thank Mickael Bourge and Mario Gomez Pacheco for their technical assistance in cytometry at the Imagerie-Gif, Plateforme de Cytométrie C.N.R.S.-I2BC, Gif-sur-Yvette, France. We are also grateful to Fatima Gaboun, for her help in realization and verification of statistical analysis, and Dr. Mohamed Ibn Tattou for plant identification and depositing of herbarium vouchers in the Scientific Institute of Rabat (S.I.R.), Morocco.
References
- Aboukhalid, K., A. Lamiri, M. Agacka-Mołdoch, T. Doroszewska, A. Douaik, M. Bakha, J. Casanova, F. Tomi, N. Machon, and C. Al Faiz. 2016. “Chemical Polymorphism of Origanum compactum Grown in all Natural Habitats in Morocco.” Chemistry and Biodiversity 13 (9): 1126–1139.10.1002/cbdv.v13.9
- Astanova, S. B. 1981. ‘‘Novye dannye o khromosomnikh chislakh nekotorykh vidov gubocvetnykh Tadzhikistana. Izv. Akad. Nauk Tadziksk. SSR, Otd.’’ [New Data on the Chromosome Numbers of Some Species of Labiatae of Tajikistan Izv. Acad. Science Tajik. SSR, Otd] Biologicheskikh Nauk 1 (82): 10–15.
- Ayyangar, K. R., and B. Vembu. 1985. ‘‘Karyo-Specific and Karyo-Generic Affiliations Amongst Mentha arvensis Benth., M. piperita L. and Origanum vulgare L.’’ Proceedings of the Indian Science Congress Association 72 (3–VI): 127.
- Azizi, A., F. Yan, and B. Honermeier. 2009. “‘Herbage Yield, Essential Oil Content and Composition of Three Oregano (Origanum vulgare L.) Populations as Affected by Soil Moisture Regimes and Nitrogen Supply.” Industrial Crops and Products 29 (3): 554–561.10.1016/j.indcrop.2008.11.001
- Bakhy, K., O. Benlhabib, A. Bighelli, J. Casanova, F. Tomi, and C. Al Faiz. 2014. “Yield and Chemical Variability of the Essential Oil Isolated from Aerial Parts of Wild Origanum compactum Benth. From Moroccan Western Rif.” American Journal of Essential Oils and Natural Products 1 (4): 9–17.
- Balim, A. G., and T. Kesercioğlu. 1998. ‘‘Doğu Akdeniz Bölgesinde yayılış gösteren bazı Origanum L. türleri üzerinde sitotaksonomik araştımalar. XIV.’’ [Cytotaxonomic Studies on Some Species of Origanum L. distributed in the Eastern Mediterranean region. XIV] Ulusal Biyoloji Kongresi 1: 277–282.
- Bastida, F., and S. Talavera. 1994. ‘‘Nǔmeros cromosomăticos de plantas occidentales, 688–695.’’ [Chromosome Numbers of Western Plants, 688-695] Anales del Jardín Botánico de Madrid 51 (2): 279–280.
- Bennett, M. D., and I. J. Leitch. 2012. Plant DNA C-values database (release 6.0, Dec. 2012). Accessed October 17, 2017 http://data.kew.org/cvalues/.
- Boscaiu, M., J. Riera, E. Estrelles, and J. Güemes. 2000. ‘‘Números cromosomáticos de plantas occidetales, 827-848.’’ [Chromosome Numbers of Western Plants, 827-848] Anales del Jardín Botánico de Madrid 58 (1): 163–164.
- Bou Dagher-Kharrat, M., N. Abdel-Samad, B. C. Douaihy, M. Bourge, A. Fridlender, S. Siljak-Yakovlev, and S. Brown. 2013. “Nuclear DNA C-Values For Biodiversity Screening: Case of the Lebanese Flora.” Plant Biosystems 147 (4): 1228–1237.10.1080/11263504.2013.861530
- Bräuchler, C., H. Meimberg, and G. Heubl. 2010. “Molecular Phylogeny of Menthinae (Lamiaceae, Nepetoideae, Mentheae) – Taxonomy, Biogeography and Conflicts.” Molecular Phylogenetics and Evolution 55 (2): 501–523.10.1016/j.ympev.2010.01.016
- Cardona, M. A. 1973. “Contribution à l’étude cytotaxonomique de la flora des Baléares I [Contribution to Cytotaxonomic Study of Flora of Balearic I].” Acta Phytotaxonomica, Barcinonensia 14: 1–20.
- Carlström, A. 1984. “New species of Alyssum, Consolida, Origanum and Umbilicus from SE Aegean Sea.” Willdenowia 14: 15–26.
- Danin, A. 1990. “Two New Species of Origanum (Labiatae) from Jordan.” Willdenowia 19: 401–405.
- Danin, A., and I. Künne. 1996. “Origanum jordanicum (Labiatae), a New Species from Jordan, and Notes on Other Species of Sect. Campanulaticalyx.” Willdenowia 25: 601–611.
- Dobignard, A., and C. Chatelain. 2010‒13. ‘Index synonymique et bibliographique de la flore d’Afrique du Nord [Synonymic and Bibliographic Index of North Africa Plants].” vol. 1-5. African Plant Database (version 3.4.0). Conservatoire et Jardin botaniques de la Ville de Genève [Museums of Geneva] and South African National Biodiversity Institute, Pretoria. http://www.ville-ge.ch/musinfo/bd/cjb/africa/.
- Duman, H., Z. Aytec, M. Ekici, E. A. Karaveliogullari, A. Donmez, and A. Duran. 1995. “Three New Species (Labiatae) from Turkey.” Flora Mediterranea 5: 221–228.
- Duman, H., K. H. C. Baser, and Z. Aytec. 1998. “Two New Species and a New Hybrid from Anatolia.” Turkish Journal of Botany 22: 51–55.
- El Oualidi, J., and T. Navarro. 2007. “Genre Origanum in Flore Pratique du Maroc [Genus Origanum in Practical Flora of Morocco].” In edited by M. Fennane, M. Ibn Tattou, A. Ouyahya, and J. El Oualidi, 476–477. Rabat, Maroc: Institut Scientifique de Rabat.
- El Oualidi, J., H. Khamar, M. Fennane, M. Ibn Tattou, S. Chauvet, and M. S. Taleb. 2012. ‘Check-list des endémiques et spécimens types de la flore vasculaire de l’Afrique du Nord [Checklist of Endemic and Typical Specimens of the Vascular Flora of North Africa]. “Document de L’Institut Scientifique N°25. Rabat: Université Mohammed V-Agdal.
- Esra, M., H. Duman, and F. Ünal. 2008. “Karyological Studies of Five Taxa of Sideritis L. (Lamiaceae) section Hesiodia Benth. from Turkey’.” Caryologia 61 (2): 115–122.10.1080/00087114.2008.10589617
- Euro+Med. 2006. “Euro+Med PlantBase - The Information Resource for Euro-Mediterranean Plant Diversity.” Accessed October 17, 2017. http://ww2.bgbm.org/EuroPlusMed/.
- Fernandes, A., and M. T. Leitão. 1984. ‘‘Contribution à l’étude cytotaxinomique des Spermatophyta du Portugal XVIII – Lamiaceae.’’ [Contribution to the Cytotaxinomic Study of Spermatophyta of Portugal XVIII-Lamiaceae] Memorias da Sociedade Broteriana. 27: 27–75.
- Feulgen, R., and H. Rossenbeck. 1924. “Mikroskopisch-chemischer Nachweis einer Nukleinsäure vom Typus der Thymonukleinsäure und die darauf beruhende elektive Färbung von Zellkernen in mikroskopischen Präparaten [Microscopic Chemical Detection of a Nucleic Acid of the Thymonucleic Acid Type and the Elective Staining of Nuclei in Microscopic Specimens Based Thereon].” Hoppe-Seyler’s Zeitschrift fur physiologische Chemie 135 (5): 203–248.10.1515/bchm2.1924.135.5-6.203
- Gill, L. S. 1981. “Chromosomal Evolution and Incidence of Polyploidy in the Canadian Labiatae.” Revue de cytologie et de biologie végétales le Botaniste 4: 331–339.
- Goldblatt, P., and D. E. Johnson, eds. 1979. Index to Plant Chromosome Numbers. St. Louis: Missouri Botanical Garden. Accessed October 17, 2017. http://www.tropicos.org/Project/IPCN/.
- Harriman, N. A. 1975. “In IOPB Chromosome Number Reports XLVIII.” Taxon 24: 367–372.
- Ietswaart, J. H. 1980. ‘A Taxonomic Revision of the genus Origanum (Labiatae).’’ In Leiden Botanical Series, vol. 4. The Hague: Leiden University Press.
- Kokkini, S. 1997. ‘Taxonomy, Diversity and Distribution of Origanum Species.’’ In Oregano. Proceedings of the IPGRI international workshop, 8-12 May 1996, CIHEAM, Valenzano, Bari, Italy, edited by S. Padulosi, 2–12. Roma: IPGRI.
- Krasnikov, A. A., and D. N. Schaulo. 1990. “Chromosome Numbers in Representatives of Some Families of Vascular Plants in the Flora of the Novosibirsk Region. II.” Botanicheskii Zhurnal. Moscow & Leningrad 75: 118–120.
- Leitch, I. J., D. E. Soltis, P. S. Soltis, and M. D. Bennett. 2005. “Evolution of DNA Amounts Across Land Plants (Embryophyta).” Annals of Botany 95 (1): 207–217. 10.1093/aob/mci014
- Lepers-Andrzejewski, S., S. Siljak-Yakovlev, S. C. Brown, M. Wong, and M. Dron. 2011. “Diversity and Dynamics of Plant Genome Size: An Example of Polysomatry from a Cytogenetic Study of Tahitian Vanilla (Vanilla ×tahitensis, Orchidaceae).” American Journal of Botany 98 (6): 986–997.10.3732/ajb.1000415
- Lepper, L. 1970. ‘Beiträger zu einer Flora des Orientes.’’ [Contributors to a Flora of the Orient] Linnaea 21: 639–663.
- Lövkvist, B., and U. M. Hultgård. 1999. “Chromosome Numbers in South Swedish Vascular Plants.” Opera Botanica 137: 1–42.
- Lukas, B., and J. Novak. 2013. “The Complete Chloroplast Genome of Origanum vulgare L. (Lamiaceae).” Gene 528 (2): 163–169.10.1016/j.gene.2013.07.026
- Lukas, B., R. Samuel, E. Mader, K. H. Başer, H. Duman, and J. Novak. 2013. “Complex Evolutionary Relationships in Origanum section Majorana (Lamiaceae).” Botanical Journal of the Linnean Society 171: 667–686.10.1111/boj.2013.171.issue-4
- Lukas, B., C. Schmiderer, and J. Novak. 2013. “Phytochemical Diversity of Origanum vulgare subsp. vulgare L. (Lamiaceae) from Austria.” Biochemical Systematics and Ecology 50: 106–113. 10.1016/j.bse.2013.03.037
- Luque, T., and Z. D. Lifante. 1991. “Chromosome Numbers of Plants Collected During Iter Mediterraneum I in the SE of Spain.” Bocconea 1: 303–364.
- Magulaev, A. V. 1984. “Cytotaxonomic Study in Some Flowering Plants of the North Caucasus.” Botanicheskii Zhurnal 69 (4): 511–517.
- Májovský, J., ed. 1978.’’Index of Chromosome Numbers of Slovakian flora (Part 6).’’ Acta Facultatis Rerum Naturalium Universitatis Comenianae. Botanica 26: 1–42.
- Marie, D., and S. C. Brown. 1993. “A Cytometric Exercise in Plant DNA Histograms, with 2C Values for 70 Species.” Biology of the Cell 78 (1): 41–51.10.1016/0248-4900(93)90113-S
- Markova, M., and V. Goranova. 1995. “Mediterranean Chromosome Number Reports 5 (435–473).” Flora Mediterranea 5: 289–317.
- Mas de Xaxars, G., T. Garnatje, J. Pellicer, S. Siljak-Yakovlev, J. Vallès, and S. Garcia. 2015. “Impact of Dysploidy and Polyploidy on the Diversification of High Mountain Artemisia (Asteraceae) and Allies.” Alpine Botany 126 (1): 35–48.
- Mehrpur, S., H. Mirzaie-Nodoushan, A. Majd, and F., Sefidkon. 2002. “Karyotypic Studies of Two Thymus Species.” Cytologia 67 (4): 343–346.10.1508/cytologia.67.343
- Montmollin, B. 1986. “Etude cytotaxonomique de la flore de la Crète. III. Nombres chromosomiques.” [Cytotaxonomic Study of the flora of Crete. III. Chromosome Numbers] Candollea 41: 431–439.
- Morales, R. 1990. ‘‘Números cromosomáticos de plantas occidentales, 582-590.’’ [Chromosome numbers of western plants, 582-590] Anales del Jardín Botánico de Madrid 47: 193–198.
- Mowforth, M. 1986. Variation in Nuclear DNA Amounts in Flowering Plants: An Ecological Analysis, PhD diss., University of Sheffield, United Kingdom.
- Pastor, J., J. C. Diosdado, C. S. Bárbara, J. Vique, and E. Pérez. 1990. ‘‘Números cromosómicos para la flora Española, 556–591.’’ [Chromosome Numbers for the Spanish Flora, 556–591] Lagascalia 15: 269–282.
- Rice, A., L. Glick, S. Abadi, M. Einhorn, N. M. Kopelman, A. Salman-Minkov, J. Mayzel, O. Chay, and I. Mayrose. 2014. “The Chromosome Counts Database (CCDB) – A Community Resource of Plant Chromosome Numbers.” New Phytologist 206 (1): 19–26.
- Siljak-Yakovlev, S., V. Stevanovic, M. Tomasevic, S. C. Brown, and B. Stevanovic. 2008. “Genome Size Variation and Polyploidy in the Resurrection Plant Genus Ramonda. Cytogeography of Living Fossils.” Environmental and Experimental Botany 62 (2): 101–112.10.1016/j.envexpbot.2007.07.017
- Skalinska, M., A. Jankun, and H. Wcislo. 1971. “Studies in Chromosome Numbers of Polish Angiosperms, Eighth Contribution.” Acta Biologica Cracoviensia Series Botanica 14: 76–77.
- Skoula, M., P. Gotsiou, G. Naxakis, and C. B. Johnson. 1999. “A Chemosystematic Investigation on the Mono- and Sesquiterpenoids in the Genus Origanum (Labiatae).” Phytochemistry 52 (4): 649–657.10.1016/S0031-9422(99)00268-X
- Stepanov, N. V., and E. N. Muratova. 1995. “Chromosome Numbers of Some Taxa of Higher Plants of Krasnoyarsk Territory.” Botanicheskii Zhurnal. Moscow & Leningrad 80 (6): 114–116.
- Strid, A., and R. Franzen. 1981. “In Chromosome Number Reports LXXIII.” Taxon 30: 829–842.
- Tahiri, B., M. Rejdali, and M. Atbib. 1998. “Contribution à l’étude caryologique de certaines espèces marocaines du genre Thymus L. (Labiatae).” [Contribution to the Karyological Study of Certain Moroccan Species of the Genus Thymus L. (Labiatae)] Flora Mediterranea 8: 41–47.
- Valdés, B., M. Rejdali, A. Achhal El Kadmiri, J. L. Jury, and J. M. Montserrat. 2002. Checklist of vascular plants of N Morocco with identification keys. (2 volume set). Catalogue des plantes vasculaires du Nord du Maroc, incluant des clés d’identification. VI. Madrid: Consejo Superior de Investigaciones Cientifeicas.
- Von Bothmer, R. 1970. “Studies in the Aegean Flora XV. Chromosome Numbers in Labiatae.” Botaniska Notiser 123: 52–60.
- Welch, B. L. 1947. “The Generalization of Student’s Problem When Several Different Population Variances are Involved.” Biometrika 34: 28–35.
- Yang, Z. J., L. Zhang, H. X. Zhao, R. W. Yang, C. B. Ding, Y. H. Zhou, and D. G. Wan. 2009. “Chromosome Numbers of Some Species of Salvia (Lamiaceae) from the Sichuan Province, China.” Nordic Journal of Botany 27 (4): 287–291.10.1111/njb.2009.27.issue-4
- Yildiz, K., and S. Gücel. 2006. “Chromosome Numbers of 16 Endemic Plant Taxa from Northern Cyprus.” Turkish Journal of Botany 30: 181–192.