?Mathematical formulae have been encoded as MathML and are displayed in this HTML version using MathJax in order to improve their display. Uncheck the box to turn MathJax off. This feature requires Javascript. Click on a formula to zoom.
?Mathematical formulae have been encoded as MathML and are displayed in this HTML version using MathJax in order to improve their display. Uncheck the box to turn MathJax off. This feature requires Javascript. Click on a formula to zoom.Abstract
Color, texture, and shape are the common features used for the retrieval systems. However, many medical images have a spot of color information. Therefore, the discriminative texture and shape features should be extracted to obtain a satisfied retrieval result. In order to increase the credibility of the retrieval process, many features can be combined to be used for medical image retrieval. Meanwhile, more features require more processing time, which will decrease the retrieval speed. In this paper, wavelet decomposition is adopted to generate different resolution images. Bag-of-feature, texture, and LBP feature are extracted from three different-level wavelet images. Finally, the similarity measure function is obtained by fusing these three types of features. Experimental results show that the proposed multi-feature fusion method can achieve a higher retrieval accuracy with an acceptable retrieval time.
1. Introduction
The medical content-based image retrieval (M-CBIR) systems have been developed and used for pathology, radiology, and clinical laboratory diagnostics [Citation1]. In these systems, some databases only contain a single kind of medical image, while others contain many kinds of medical images. Radiology images not only have a large number but also play an important role in auxiliary diagnosis. Therefore, the radiation image retrieval system has become a hot research area [Citation2].
Since it is easy to identify color and texture in the pathological image, CBIR is often used in the research of retrieval system of pathological images. A computer-aided diagnosis system was proposed for pigmented skin lesions and multiple classifier systems were used for melanoma diagnosis [Citation3]. To retrieve the medical image, the traditional global features have been widely used including color features, texture features, and shape features. Compared with text retrieval, image retrieval often constructs visual words by utilizing all kinds of features [Citation4]. ASSERT is a retrieval system for lung CT image. In the retrieval algorithm of ASSERT, lesion feature is characterized by combining texture with shape information [Citation5]. Cardiac CT images are important parts of the medical database, and related retrieval methods have been formed by taking advantage of the heart shape. A contour and texture based image retrieval technology has been put forward and applied to the liver image database [Citation6]. Patch-based features have been applied to X-ray medical image retrieval, while scale invariant feature transform (SIFT) has been selected as well as a new feature for medical image retrieval.
Since wavelet transformation can extend an image into different size and obtain idiographic feature information, it is also introduced to medical image retrieval. By combining Gabor filter and Euclidean distance, wavelet transform has achieved better performance on image retrieval. When features have been selected, image retrieval needs good methods to classify these features in order to obtain the best result similar to query image [Citation7]. Support vector machine is in common use for retrieval in medical image database including X-Rays, MRI, and CT. IRMA is an efficient image retrieval system using feature fusion and support vector machine [Citation8].
To improve the performance of CBIR for a medical image, machine learning has been used in pre-filtering image and statistical similarity has been used in the matching of multi-feature between query image and database [Citation9]. UMLS is a successful medical image retrieval system which has a structured learning framework and modular design based on support vector machine. To find out the possible lesion region in brain images, domain knowledge has been used in retrieving the image sequence. A boosting framework has been proposed to improve the performance of medical image retrieval. Evaluation results show that this method has a high retrieval accuracy with a low computational cost [Citation10].
In this paper, a multi-feature fusion method was proposed for medical image retrieval based on wavelet transform and bag-of-features. The remaining parts of this paper are organized as follows: Framework of the proposed method is founded in Section 2. In Section 3, mathematical expression and execution details of the proposed method are listed. Experiments are shown in Section 4 and conclusion is listed in Section 5.
2. Retrieval framework of the proposed method
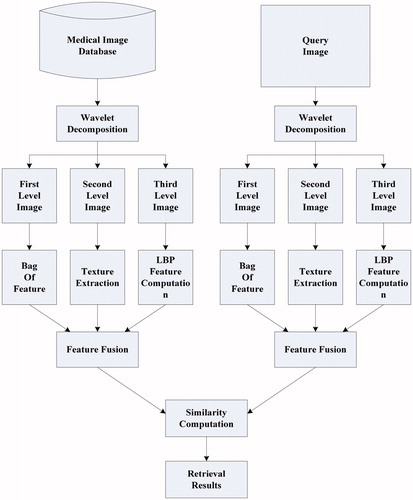
The retrieval framework of the proposed method is shown as in
From , we can see that this method will be implemented by seven steps:
Different resolution images are obtained by using wavelet decomposition.
Gray-based bag-of-features are computed according to first-level resolution image.
Texture features are extracted from second-level resolution image.
LBP features are computed by utilizing third-level resolution image.
Retrieval feature is obtained by fusing bag-of-features, texture feature, and LBP feature.
Comparing retrieval features in between query image and image of medical database.
Retrieval results are output by the order of similarity.
3. Multi-feature fusion retrieval method based on wavelet and bag-of-feature
3.1. Feature selection by hash coding
In order to increase the reliability of medical image retrieval, more image features should be extracted. However, the medical image is generally large so that more time will be expended for computing these features on the original image. For this reason, wavelet decomposition was introduced to obtain multi-level resolution expression of the original image. Then, different features would be obtained on different level resolution image.
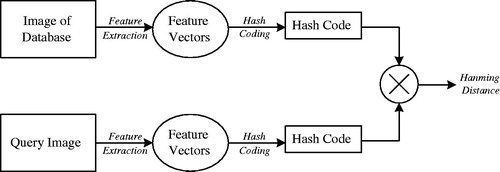
In order to get more reliable features of medical image retrieval, a certain number of images are randomly selected from the database as training images for feature extraction. The extracted features include color features, bag-of-features, texture features, shape features, LBP local features, and so on. The feature vectors of different dimensions are formed, and hash coding is generated by using these vectors as the input of hash algorithm. And then the Hamming distance is compared to determine the effectiveness of various features. It is shown as shown in .
Finally, three features are proved to be more effective for image retrieval in the medical image library. Bag-of-feature can better describe the local grayscale statistical information of medical images. Texture feature is the most abundant expression in medical image content. LBP feature has grayscale invariance and rotation invariance. It is also very important for medical image analysis.
3.2. Wavelet decomposition
The process of wavelet decomposition can be described as .
Where is the original medical image,
,
,
,
are the results after one-time wavelet decomposition, and
,
,
,
are the results after k-time wavelet decomposition.
From , we can find ,
,
,
are the same-size sub-image and express partial information of
. Here,
has more information than other three components and it can be taken as the most powerful expression of
. From the image size,
is a quarter of
. When the same computation is implemented on
, there will be less time cost than on
.
If wavelet decomposition is taken on an image with the size , the process can be described in a set of mathematical formulae as follows:
(1)
(1)
Here,
.
(2)
(2)
Here,
.
(3)
(3)
Here,
.
(4)
(4)
Here,
.
and
represents the low-pass filter and high-pass filter after the wavelet decomposition respectively, 2↓1(1↓2) represents the sampling along the column/row,
represents the level of the wavelet decomposition.
3.3. Bag-of-feature computation
Original image can be looked on as image of wavelet decomposition, and it includes the true information of disease. So
image plays the most role of extracting feature during medical image retrieval. Because
image has too large size, a long time will be cost for computing retrieval feature on it. Therefore, it is a good idea to segment
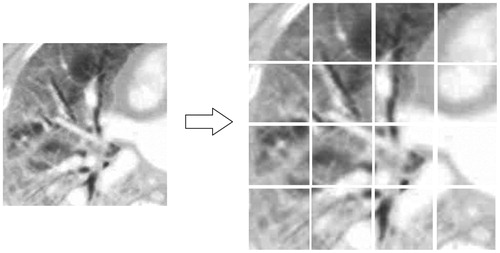
image to several small images. And these images are so small that retrieval feature can be computed via less time. The information of every bag-of-feature obtained from small image will be used in forming a feature set which can express the true information of original image.A thoracic medical image (pixels size: 256 × 256) had been segmented 16 small images shown as .
The number of small images should be determined by the size of original image. In , the size of thoracic image is 256 × 256 and the number of small image is 16. If the size of original image is bigger, the number of small image is also increased.
After segmentation, the information of all small images can be described with a corresponding matrix just like formula (5).
(5)
(5)
Where, is a feature clustering function which can express corresponding small image. In this paper, general gray-statistic function had been selected as a feature computation formula. It is shown as below:
(6)
(6)
Where,
and
are the width and height of the small image, (
,
) is the coordinate of the center pixel of small image, (
,
) is the coordinate of the arbitrary pixel of small image.
can be computed as formula (7).
(7)
(7)
3.4. Texture feature computation
Texture feature is more important to medical image retrieval because that color information is not rich. In this paper, computation about texture feature is implemented on image. In order to obtain texture feature, gray level co-occurrence matrix (GLCM) is used. GLCM characterizes the correlation between any two gray level of the medical image. As a common means of describing image texture, GLCM can be normalized to
. GLCM-based texture feature is generally divided into four kinds:
3.4.1. Energy
Energy describes the uniformity of gray distribution about an image. If data is more concentrated near the main diagonal, we can consider that gray distribution is relatively symmetrical. In this case, big energy value shows that image texture is coarse. Energy can be computed according to formula (8).
(8)
(8)
3.4.2. Entropy
Entropy is a measure of the amount of information. If entropy value is large, we can consider that there is more texture information. Entropy can be computed according to formula (9).
(9)
(9)
3.4.3. Contrast
Contrast is a measure of the clarity of image texture. If texture feature is more clear, the value of contrast should be large. Contrast can be computed according to formula (10).
(10)
(10)
3.4.4. Correlation
Correlation is the gray similarity about the row or column elements of GLCM. Correlation can be computed according to formula (11).
(11)
(11)
Where,
,
,
, and
are mean and variance of GCLM. They can be computed by using formula (12).
(12)
(12)
In this paper, the whole texture feature is made up of energy, entropy, contrast, and correlation, and it is just like formula (13).
(13)
(13)
From formula (13), energy, entropy, contrast, and correlation is considered to have the same impact on the whole texture feature.
3.5. LBP feature computation
LBP (local binary patterns) feature is also an important feature which can be used in expressing image texture. However, computing time of LBP feature will increase significantly when calculation neighborhood increases. In this paper, LBP feature is computed on image in order to spend save time.
Take a local image of 3 × 3 pixels for an example, LBP feature can be calculated as follows:
At first, center pixel is selected as the pixel to be processed and its gray value is selected as a threshold. Other pixels can be binarized by comparing their gray values with this threshold. This process is shown as formula (14).
(14)
(14)
Where,
is the gray value of center pixel,
is other pixel in local image.
After binarization, all will be combined into an 8-bit binary number and the decimal number of a binary number is LBP feature value of the center pixel. This calculation process is just like formula (15).
(15)
(15)
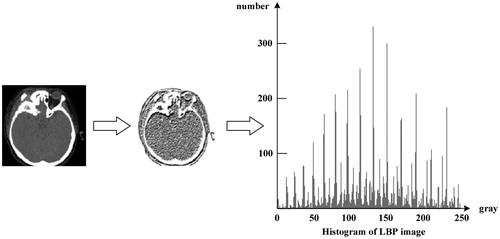
An LBP image can be obtained when LBP feature calculation is carried out over the whole image and every pixel is replaced by LBP value. Because texture feature is more legible in LBP image, LBP value has been selected as important retrieval feature. LBP results of a medical image and its histogram are shown as .
In fact, LBP feature computation can be extended to the neighborhood of arbitrary size. Because this process is carried out on image in this paper, the size of 3 × 3 pixels neighborhood is enough.
3.6. Feature fusion and similarity measurement
Retrieval results can be obtained by comparing the similarity between query image and database image. So an efficient similarity measure function is very important to image retrieval. In this paper, bag-of-feature, texture feature, and LBP feature should have a proper position in similarity measure function which is shown as formula (16).
(16)
(16)
Where,
,
, and
represents the component of bag-of-feature, texture feature, and LBP feature,
,
, and
is the weight of
,
, and
.
can be computed according to formula (17).
(17)
(17)
Where
represents the image in medical image database,
represents query image.
can be calculated by the formula (18).
(18)
(18)
The computation of is shown as formula (19).
(19)
(19)
Where, and
represents the ith probability of LBP histogram of query image and medical image of database.
Since three features were calculated on different levels of wavelet image, they should have a different effect to similarity measure function. As an expression of original image information, bag-of-feature should occupy the most important position. LBP feature is more reliable than general image texture, but information of is not rich. Therefore,
,
, and
is given by 0.4, 0.3, and 0.3.
4. Experiments and analysis
In order to test the performance of the retrieval algorithm proposed in this paper, an experimental system had been built for medical images and some experiments had been carried out. The medical image data set is selected from the CT Department of Harbin Medical University, including different CT images of all parts of the human body. Because of the confidentiality of the dataset, only a small amount of results can be shown here. The primary processor of the system is a computer which has a 2.5 GHz two core Intel CPU and an 8 G RAM. The image retrieval software is coded with C++ language, and key retrieval technology is based on multi-feature fusion algorithm proposed in this paper. The retrieval database has 1000 medical images including head CT, chest CT, pelvic CT, and spine CT. In the experiment, the key parameters are as follows: the bit number of hash codes is 32; layer number of wavelet decomposition is 2; Bags of image are 16; LBP template size is 3*3; fusion parameters ,
, and
of three features is given by 0.4, 0.3, and 0.3.
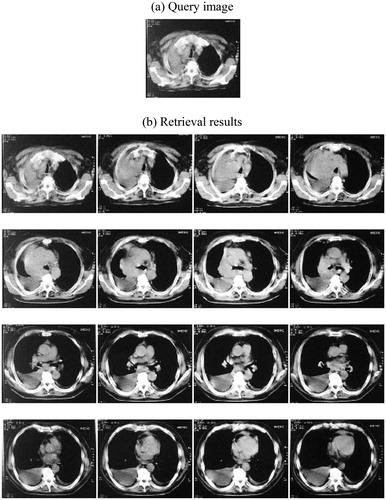
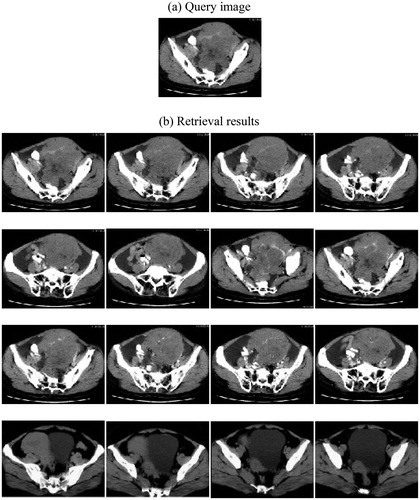
Three experimental results are shown as and .
From and , we can see that head CT, chest CT, and pelvic CT are selected as query image and retrieval results are placed order according to similarity. In fact, there are much more kinds of medical images. Limited by the capability of this paper, many experimental results can not be shown.
In order to compare retrieval performance between the proposed method and others, we had carried out image retrieval by using three different methods.
The first method is the combination of wavelet transform and energy extraction (WAVE) in the literature [Citation7]. The second method is the multi-feature SVM fusion method (SVM) in the literature [Citation9], and the third method is the multi-feature fusion method (OUR) proposed in this paper.
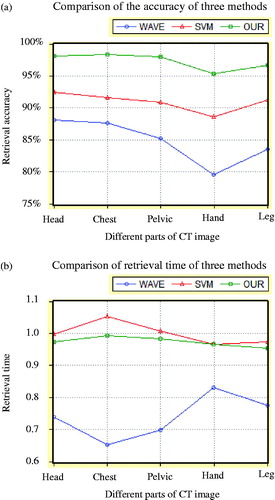
Comparison of the accuracy and retrieval time of the three methods is shown in .
From , we can see that the proposed method has the highest retrieval accuracy because multi-feature was fused in it. Moreover, retrieval time of the proposed method is not significantly increased because different feature had been computed on different wavelet- level image.
Although our method takes a longer time than the single feature method in the literature [Citation7], our method is very close with the method of multi-feature fusion in the literature [Citation9]. According to the accuracy of comprehensive retrieval, our method is obviously more ideal.
5. Conclusion
Since a single feature can bring on false retrieval results, much more reliable feature should be used for medical image retrieval. However, retrieval time will increase evidently when many features were computed during retrieval. In this paper, a multi-feature fusion method was proposed for medical image retrieval. At first, wavelet decomposition was used in obtaining multi-resolution expression of the original image. Then, bag-of-feature, texture-feature, LBP feature was computed on different resolution images. These three features were fused when retrieval similarity was measured between the query image and image of the medical image database. Experimental results show that the proposed multi-feature fusion method has a higher retrieval accuracy and its retrieval time is not added comparing with those methods using a single feature.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Li Z, Zhang X, Muller H, et al. Large-scale retrieval for medical image analytics: a comprehensive review[J]. Med Image Anal. 2018;43:66–72.
- MM, Rahman S, Antani GR. Thoma A classification-driven similarity matching framework for retrieval of biomedical images[C]. 11th ACM International Conference on Multimedia Information Retrieval, Philadelphia, USA; 2010. p.147–154.
- Shi X, Xing F, Xu K, et al. Supervised graph hashing for histopathology image retrieval and classification[J]. Med Image Anal. 2017;42:117–126.
- Bai X, Yang X, Latechi J. Learning context sensitive shape similarity by graph transduction[J]. IEEE Tran Pattern Anal Mach Intell 2010;32:861–874.
- Avni U, Greenspan H, Sharon M. X-ray image categorization and retrieval using patch-based visual words representation. Vol. 1. In Proceedings of IEEE International Symposium on Biomedical Imaging: From Nano to Macro, Rotterdam, Netherlands; 2009. p. 350–353.
- Nowakova J. M. Prilepok, V. Snasel . Medical image retrieval using vector quantization and fuzzys-tree[J]. J Med Sys 2017;41:18.
- K, Rajakumar S. Muttan. Medical image retrieval using energy efficient wavelet transform[C]. Second International Conference on Computing, Communication and Networking Technologies, Wuzhen, China; 2010. p. 1–5.
- Sasi Kumar M, Kumaraswamy YS. An improved support vector machine kernel for medical image retrieval system[C]. Proceedings of the International Conference on Pattern Recognition, Informatics and Medical Engineering; 2012. p. 257–260.
- Yonggang H, Jun Z, Yongwang Z, et al. Medical image retrieval with query-dependent feature fusion based on one-class SVM[C]. 13th IEEE International Conference on Computational Science and Engineering, Faro, Portugal; 2010. p. 176–183.
- Liu Y, Rahul S, Steven CH. A boosting framework for visuality preserving distance metric learning and its application to medical image retrieval[J]. IEEE Trans Pattern Anal Mach Intell 2010;32(1):30–44.