?Mathematical formulae have been encoded as MathML and are displayed in this HTML version using MathJax in order to improve their display. Uncheck the box to turn MathJax off. This feature requires Javascript. Click on a formula to zoom.
?Mathematical formulae have been encoded as MathML and are displayed in this HTML version using MathJax in order to improve their display. Uncheck the box to turn MathJax off. This feature requires Javascript. Click on a formula to zoom.Abstract
Liver segmentation from CT is regarded as a prerequisite for computer-assisted clinical applications. However, automatic liver segmentation technology still faces challenges due to the variable shapes and low contrast. In this paper, a patient-specific probabilistic atlas (PA)-based method combing modified distance regularized level set for liver segmentation is proposed. Firstly, the similarities between training atlases and testing patient image are calculated, resulting in a series of weighted atlas, which are used to generate the patient-specific PA. Then, a most likely liver region (MLLR) can be determined based on the patient-specific PA. Finally, the refinement is performed by the modified distance regularized level set model, which takes advantage of both edge and region information as balloon force. We evaluated our proposed scheme based on 35 public datasets, and experimental result shows that the proposed method can be deployed for robust and precise liver segmentation, to replace the tedious and time-consuming manual method.
1. Introduction
Abdominal liver segmentation is a fundamental step in liver quantitative analysis. However, due to the low contrast and variable shape, robust and accurate liver segmentation is still challenging, and it is often insufficient to segment the liver from CT using traditional segmentation techniques. Currently, varieties of approaches have been proposed for liver segmentation [Citation1]. These studies segment liver either based on pure image information or based on model.
For pure image information-based approaches, Ruskó et al. [Citation2] first used the 3 D region growing to initialize the liver contour, and then utilized post-processing techniques, such as separating the heart, removing the inferior vena cava to refine the result. However, the grayscale-based liver segmentation results are often not satisfactory, since the liver and the other adjacent organs have similar gray values in CT.
In the case of clustering or classification-based approaches, novel fuzzy clustering methods proposed by Jiang et al. (e.g. Co-FCM [Citation3] and KL-TFCM [Citation4]), have significantly improved the performance of segmentation. Li et al. [Citation5] proposed to integrate the both probabilistic shapes and global intensities into the Bayes classification frame, to achieve simultaneous segmentation of multiple organs from the abdominal CT. Meanwhile, multi-feature classifier based multi-layer liver segmentation method is explored by Selver et al. [Citation6].
Liver segmentation is also performed by the snake model. Lu et al. [Citation7] proposed a semi-automatic snake-based method. The method first initializes the snake model by the existing segmentation result (3 D mesh), and in the subsequent deformation process, the internal force, the external force and the newly introduced attractive force are used together to determine the action of each node on the grid surface.
Compared with the snake model-based approaches, the level se is another type of deformation model-based method, which is more commonly employed for liver segmentation. Generally, the level set-based methods tend to combine with other information (e.g. statistical prior knowledge, local or global image features) to improve the accuracy and reduce the computational complexity. Yang et al. [Citation8] proposed a semi-automatic segmentation method for liver CT, which uses level set to achieve refinement. Kohlberger et al. [Citation9] presented a level set approach utilizing local shape and appearance prior model, in which the local intensity statistics and local curvature information are integrated, while tracking the evolution of each level set point, and thus achieves a satisfactory result.
Since the gray scales of the organs in the abdominal CT are similar, integrate the shape prior information may significantly improve the precision of the segmentation. Zhang et al. [Citation10] utilized the statistical shape model combining with the optimal surface acquisition to achieve the automatic liver segmentation. Tomoshige et al. [Citation11] proposed a method based on conditional statistical shape model. The method first uses the maximum a posteriori (MAP) estimation to extract conditional features, and then by employing the level set to obtain a liver shape prior estimation. Finally, an accurate refinement of the liver contour was achieved by employing graph cut method.
Atlas-based method is another popular approach for robust liver segmentation. Zhou et al. [Citation12] first used liver probabilistic atlas for automatic liver segmentation in CT. The probabilistic atlas constructed in this method provides two kinds of liver information: liver position and liver grayscale, and experiment shows that their atlas-based method can reduce the calculation cost of liver segmentation process, and can significantly improve the robustness.
Although the accuracy of liver segmentation is greatly improved by the presented methods, the existing methods are still difficult to meet the requirements in clinical practice. Aiming at the low contrast between organs in the abdominal CT image, organ pathology and the difference of organ shape between individuals, this paper proposes an automatic liver segmentation method based on the combination of probabilistic atlas and modified distance regularized level set model.
2. Method
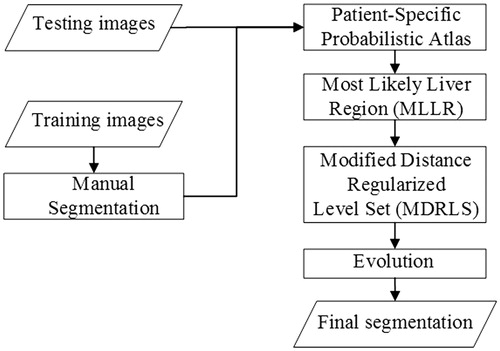
In this section, the proposed segmentation framework will be described in detail. The flowchart is depicted in .
2.1. Denoising
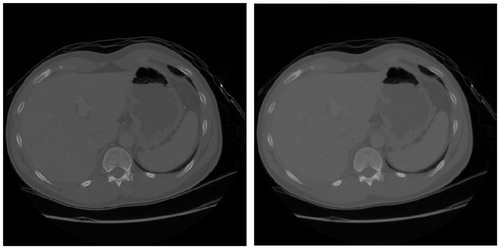
For noise reduction, this paper employs the Catte_PM anisotropic diffusion filter [Citation13] to denoise the liver CT image. shows a result of filtering the liver CT image using the Catte_PM model. As seen in the figure, the Catte_PM model can not only smooth the inner region of the organ, but also effectively preserve the edge information between organs.
2.2. Patient-Specific probabilistic atlas
For a given test dataset (patient image), the Patient-Specific PA can be dynamically generated by the following steps. First, the image similarity of all the maps and the test set is calculated, and the atlases are arranged in descending order in accordance with the obtained image similarity. The atlas is composed of N training CT datasets with corresponding “golden standards”. The similarity between the test patient and training atlas can be obtained according to the Normalized Cross Correlation (NCC) [Citation14] method, and by employing the method proposed by Chu [Citation15], a weighted probabilistic atlas is calculated.
2.3. Most likely liver region (MLLR)
After constructing the weighted PA of the testing image, the PA is then thresholded, and a binarized liver parenchymal region of interest (ROI) is generated, which is called the most likely liver region (MLLR), as shown in . In order to minimize the estimation error of the MLLR, a morphological dilation operation is performed after binarization, in which spherical structural elements are used. The resulting segmentation is then assigned as the initial contour for the following level set refinement.
2.4. Modified distance regularized level set
We descript a Modified Distance Regularized Level Set-based refinement method, in which the evolution is driven by balloon forces of edge and region information, and thereby the active contour can evolve to the edge of the liver with sufficient balloon force or sufficient number of iterations. Moreover, edge leaks do not easily occur, resulting in more accurate results.
The basic edge-based Distance Regularized Level Set model utilized in this paper is proposed by Li [Citation16]. Given level set equation φ: image domain Ω→R, and an energy function ε(φ) can be defined as follows
(1)
(1)
where μ > 0. Dp(φ) is a penalty term, which is aimed to avoid the re-initialization of the level set, and it is defined as follows:
(2)
(2)
(3)
(3)
where p(|▽φ|) is a double-well potential function. The main role of this function is to allow the binary Heaviside function to be directly initialized for the level set, and the function εext(φ) is an external force that can be defined according to the edge or the regional information to drive the evolution of the level set.
An edge-based DRLSE level set was constructed by adding the edge information εext(φ) of image. Define the edge indication function g:
(4)
(4)
where Gσ represents the Gaussian kernel function with variance σ, and εext(φ) is defined as follows:
(5)
(5)
where Ω is the image domain, λ andαare both constant coefficients, δis the Dirac function and H represents the Heaviside function, Lg(φ) represents the weighted length of the curve and Ag(φ) is the evolution of the balloon force and control curve.
In our modified level set model, a new balloon force based on regional information (Signed Pressure Force, SPF)proposed by Zhang [Citation17] is combined into edge-based DRLSE, and SPF(I(x)) is defined as follows,
(6)
(6)
in which c1, c2 are the average gray-level constants inside and outside the curve, which can be obtained by solving the C-V model [Citation18]. As the SPF(I(x)) is obtained, it can be incorporated into the balloon force of EquationEquation (5)
(5)
(5) to generate a new external energy functional equation:
(7)
(7)
Consequently, the energy functional of the proposed modified level set can be represented as:
(8)
(8)
2.5. Evolution
Assume I0 denotes the initial contour binary image of the live. Due to the existence of the dual-well function p(|▽φ|) in the DRLSE framework, by using binary step function, the level set the function G0 can be initialized directly, and thus simplify the initialization of SPF. The initialization equation is as follows:
(9)
(9)
where ω is a variable parameter for controlling the width of SDF. Since the image domain is discretely gridded, for the symbol distance function, there is at least one grid point on both sides of the zero level set contour. Therefore, the width of the symbol distance function is at least two [Citation16].
Through the evolution of the initialized level set, the final refinement of liver segmentation is then achieved.
3. Experiment
In the training phase, we use 20 liver contrast-enhanced CT training datasets with corresponding segmentations, which are from MICCAI 2007 (www.sliver07.org). For the training dataset, the pixel size is: 0.55 × 0.55 − 0.80 × 0.80 mm, and the layer spacing is: 1.0-3.0 mm.
In the testing phase, 15 test datasets were used to validate the accuracy of the presented method. The test dataset and its corresponding “gold standard” segmentation results are from 3 D-IRCADb (https://www.ircad.fr/research/3dircadb/). For the test dataset, the pixel size is: 0.56 × 0.56 − 0.87 × 0.87 mm, with layer spacing: 1.0-4.0 mm. Each slice in all datasets contains flat pixels of 512 × 512.
In the experiments, two comparative studies are mainly implemented. One is the comparison on segmentation errors using the proposed patient-specific PA and standard PA, respectively, and another is a comparison on segmentation errors between our proposed method and two other methods.
The experiment is implemented with MATLAB R2010a, Intel i7-4770, 16GB RAM, and in order to evaluate the accuracy, 5 metrics were chosen in this research: VOE, RVD, ASD, RMSD, and MSD [Citation1].
4. Result
According to the above five metrics, gives the evaluation details by our proposed method on 15 3Dircadb datasets, in which the golden standard was provided by radiologist.
Table 1. Segmentation errors of our method by five metrics.
To prove the superiority of patient-specific probabilistic atlas compared to standard atlas in our research, a comparative experiment is also implemented on the 15 3Dircadb datasets. The result is shown in . In addition, a comparison between the proposed method and two other approaches is shown in .
Table 2. Comparisons between standard atlas and our patient-specific PA on 15 3Dircadb datasets.
Table 3. Comparisons with other methods with 3Dircadb datasets.
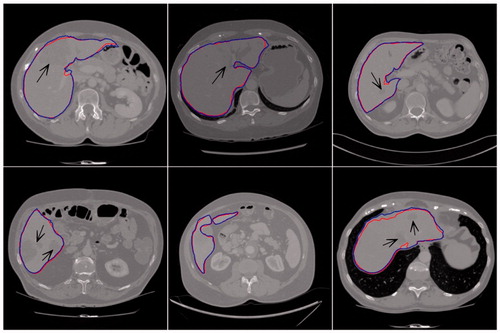
shows partial result of the proposed method (dark thick line) and manual segmentation (light thin line) on some representative slices. The black arrows indicated the liver tumor area. Comparing the results of our proposed method with ground truth, we can find that, even for slices containing tumors, our proposed method can still achieve accurate segmentation, with strong robustness.
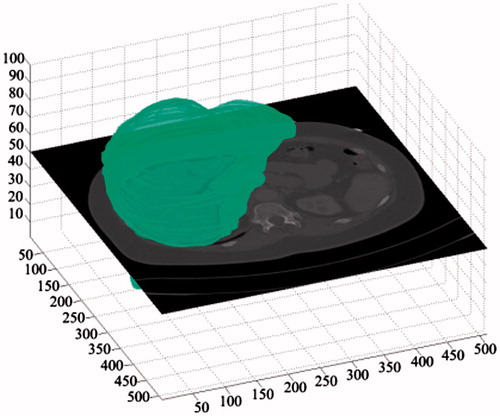
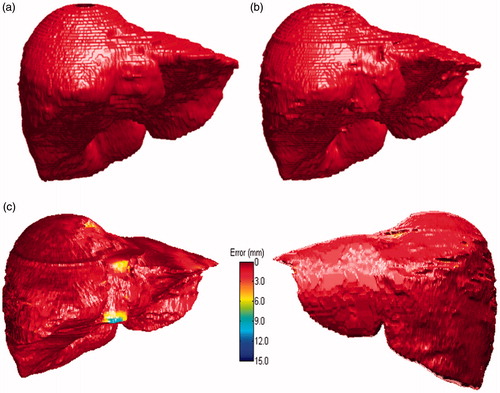
For a same segmentation, the comparative 3D visualizations using our proposed method and ground truth were depicted in ), respectively. Based on the average ASD error, a 3 D visualizations of error between our result and ground truth result is also provided in , in which, the ASD error was 1.41 mm.
5. Discussion
It can be seen from that, patient-specific PA obtained a higher accuracy on the five metrics. The main reason is that, hepatic diseases may affected some liver tissues and change its shape, meanwhile possible shape variations may also be lost while constructing such an atlas by standard atlas. To overcome these problems, standard atlas often requires a large-scale training datasets [Citation21], while in this paper the weighted patient-specific probabilistic atlas is proposed based on the similarity between target testing image and multiple clusters, so that the obtained initial shape is much more closer to the shape and position of the testing image of the patient, by making full use of limited training samples.
From the , it can be seen that, our method outperforms the other two methods on VOE, ASD, RMSD and MSD, however, a worse result is obtained on RVD. The main reason for that is, although MLLR could retain possible liver contours to the greatest extent, it also causes over-segmentation, which inevitably results in a bigger RVD error. Moreover, an attempt was also made on liver image with tumor volume more than 50%, a poor MLLR was obtained by our proposed method, and resulting in a large segmentation error in the post fine-tune processing, which indicates the limitation of our proposed method when deployed on liver CT with larger tumors. In addition, the average running time for one scan was less than 7 minutes, which was mainly exhausted on solving the minimization problem of level set.
Furthermore, to evaluate whether the difference of segmentation accuracy between the proposed method and the other two methods was statistically significant, the paired t-test was conducted, which are based on two major metrics: VOE and ASD. As a result, our proposed method outperforms the other two technologies with statistically significant differences on VOE and ASD (p < 0.05).
6. Conclusion
In this paper, a Modified Distance Regularized Level Set-based liver segmentation method is proposed using patient-specific probabilistic atlas from abdominal CT. To some challenging clinical problems, the applicability of the proposed scheme is demonstrated, and results in good performance on both quantitative and qualitative analyses. Experiment shows that the proposed scheme can delineate liver boundaries that have a level similar to those obtained manually, and with good robustness.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Heimann T, Van Ginneken B, Styner MA, et al. Comparison and evaluation of methods for liver segmentation from ct datasets. IEEE Trans Med Imaging. 2009;28:1251–1265.
- Ruskó L, Bekes G, Fidrich M. Automatic segmentation of the liver from multi-and single-phase contrast-enhanced ct images. Med Image Anal. 2009;13:871–882.
- Jiang Y, Chung FL, Wang S, et al. Collaborative fuzzy clustering from multiple weighted views. IEEE Trans Cybern. 2015;45:688–701.
- Qian P, Zhao K, Jiang Y, et al. Knowledge-leveraged transfer fuzzy c-means for texture image segmentation with self-adaptive cluster prototype matching. Knowl-Based Syst. 2017;130:33–50.
- Li C, Wang X, Li J, et al. Joint probabilistic model of shape and intensity for multiple abdominal organ segmentation from volumetric ct images. IEEE J Biome Health Inf. 2013;17:92–102.
- Selver MA. Segmentation of abdominal organs from ct using a multi-level, hierarchical neural network strategy. Comput Meth Prog Bio. 2014;113:830–852.
- Lu J, Shi L, Deng M, et al. An interactive approach to liver segmentation in ct based on deformable model integrated with attractor force. In: Machine Learning and Cybernetics (ICMLC), 2011 International Conference on; Vol. 4; IEEE; 2011. p. 1660–1665.
- Yang X, Yu HC, Choi Y, et al. A hybrid semi-automatic method for liver segmentation based on level-set methods using multiple seed points. Comput Methods Programs Biomed. 2014;113:69–79.
- Kohlberger T, Uzunba ¸ s MG, Alvino C, et al. Organ segmentation with level sets using local shape and appearance priors. In: International conference on medical image computing and computer-assisted intervention. Berlin, (Germany): Springer; 2009. p. 34–42.
- Zhang X, Tian J, Deng K, et al. Automatic liver segmentation using a statistical shape model with optimal surface detection. IEEE Trans Biomed Eng. 2010;57:2622–2626.
- Tomoshige S, Oost E, Shimizu A, et al. A conditional statistical shape model with integrated error estimation of the conditions; application to liver segmentation in non-contrast ct images. Med Image Anal. 2014;18:130–143.
- Zhou X, Kitagawa T, Hara T, et al. Constructing a probabilistic model for automated liver region segmentation using non-contrast x-ray torso ct images. In: International conference on medical image computing and computer-assisted intervention. Berlin, (Germany): Springer; 2006. p. 856–863.
- Catt ´ e F, Lions PL, Morel JM, et al. Image selective smoothing and edge detection by nonlinear diffusion. SIAM J Numer Anal. 1992;29:182–193.
- Oda M, Nakaoka T, Kitasaka T, et al. Organ segmentation from 3d abdominal ct images based on atlas selection and graph cut. In: International MICCAI workshop on computational and clinical challenges in abdominal imaging. Berlin, (Germany): Springer; 2011. p. 181–188.
- Chu C, Oda M, Kitasaka T, et al. Multi-organ segmentation from 3d abdominal ct images using patient-specific weighted-probabilistic atlas. In: Medical Imaging 2013: Image Processing; Vol. 8669; International Society for Optics and Photonics; 2013. p. 86693Y.
- Li C, Xu C, Gui C, et al. Distance regularized level set evolution and its application to image segmentation. IEEE T Image Process. 2010;19:3243.
- Zhang K, Zhang L, Song H, et al. Active contours with selective local or global segmentation: a new formulation and level set method. Image Vis Comput. 2010;28:668–676.
- Chan TF, Sandberg BY, Vese LA. Active contours without edges for vector-valued images. J Vis Commun Image R. 2000;11:130–141.
- Li G, Chen X, Shi F, et al. Automatic liver segmentation based on shape constraints and deformable graph cut in ct images. IEEE T Image Process. 2015;24:5315–5329.
- Erdt M, Steger S, Kirschner M, et al. Fast automatic liver segmentation combining learned shape priors with observed shape deviation. In: International Symposium on Computer-Based Medical Systems; IEEE; 2010. p. 249–254.
- Linguraru MG, Sandberg JK, Li Z, et al. Atlas-based automated segmentation of spleen and liver using adaptive enhancement estimation In: international conference on medical image computing and computer-assisted intervention. Berlin, (Germany): Springer; 2009. p. 1001–1008.