Abstract
The Mediterranean Sea is an important regional management unit (RMU) for the conservation of the loggerhead turtle (Caretta caretta) because it hosts both nesting and foraging grounds for this species. In this study, attention was focused on an important foraging ground located on the southern Italian coast of the Adriatic Sea, the Gulf of Manfredonia. In this area, C. caretta is threatened by a high level of anthropogenic pressure, especially caused by fishing activities. To understand the genetic composition of the Southwestern Adriatic feeding ground and to highlight which rookeries could be the most impacted by human activities in this area, the mitochondrial haplotype of 239 individuals stranded or incidentally caught in fishing gears from 2008 to 2013 was determined. A mixed stock analysis (MSA) identified the Mediterranean rookeries as the most important contributors to this feeding ground, with contributions also from Atlantic rookeries of Florida and Gulf of Mexico. Finally, the identification of two “orphan haplotypes” and the recovery of three hatchlings on the Vieste beach draw attention to the need for better identification and genetic characterisation of new nesting sites within the Mediterranean Basin.
Introduction
The Mediterranean Sea is a semi-enclosed basin that represents 0.82% of the global oceanic surface and exceptionally hosts more than 17,000 marine species with a high degree of endemism (Coll et al. Citation2010). This makes the Mediterranean Sea one of the most important marine biodiversity hotspots in the world (Myers et al. Citation2000). However, this species richness is threatened by increasing anthropogenic pressure (Cuttelod et al. Citation2008; Claudet & Fraschetti Citation2010; Coll et al. Citation2010, Citation2012). “Bycatch”, the incidental capture of non-target species during fishing activities, is a critical threat to several long-lived and slow-maturing species and poses a global problem (Lewison et al. Citation2004, Citation2014). In this context, the Mediterranean Sea, together with the Southwest Atlantic Ocean and the Eastern Pacific Ocean, is an important hotspot for sea turtle bycatch (Lewison et al. Citation2014).
Three species of sea turtles are reported for the Mediterranean Basin: the loggerhead turtle Caretta caretta (Linnaeus, 1758), the green turtle Chelonia mydas (Linnaeus, 1758) and the leatherback turtle Dermochelys coriacea (Vandelli, 1761). Among them, Caretta caretta is the most abundant sea turtle inhabiting and nesting in the Mediterranean Sea (Broderick et al. Citation2002; Casale & Margaritoulis Citation2010). Accidental and intentional fishing (ca. 132,000 captures of sea turtles per year with 44,000 deaths; Casale Citation2011), collision with boats, pollutants, debris ingestion and habitat degradation (mainly of the nesting sites) represent the major threats to marine turtle conservation and are the main reasons for the general decline of populations in the Mediterranean Sea (Margaritoulis et al. Citation2003; Casale & Margaritoulis Citation2010).
The vulnerability of C. caretta is increased by a complex life cycle during which this species migrates through several differently impacted areas. The original developmental pattern (Bolten Citation2003) describes a first phase during which hatchlings leave natal beaches and reach oceanic areas where they start to feed on epipelagic preys. A second developmental step follows the oceanic phase. During this stage, juveniles return to neritic areas, change their feeding habits to a benthic diet and reach sexual maturity. Finally, at the time of reproduction, mature loggerhead turtles undertake a reproductive migration during which females return to their natal beaches to nest. This phylopatric behaviour is called “natal homing” (Bowen et al. Citation1994). However, some exceptions to the life cycle described above have recently been observed, highlighting the possibility of high behavioural plasticity of sea turtles (Luschi & Casale Citation2014).
The capability of C. caretta to accomplish long migrations towards foraging or nesting grounds, the fidelity of females to the nesting sites and the resulting population structure were demonstrated using different approaches such as capture–mark–recapture studies (Casale et al. Citation2007; Revelles et al. Citation2008; Moncada et al. Citation2010), satellite telemetry (Bentivegna Citation2002; Girard et al. Citation2009; Marcovaldi et al. Citation2010; Casale et al. Citation2013) and maternally inherited molecular markers (e.g. mitochondrial DNA – mtDNA henceforth; Encalada et al. Citation1998; Hatase et al. Citation2002; Carreras et al. Citation2007; Shamblin et al. Citation2012; Clusa et al. Citation2013). Specifically, the use of mtDNA has allowed the identification of genetically separate rookeries, characterised by unique haplotypes or specific haplotype frequencies. Differences in the frequency of mtDNA haplotypes between distinct rookeries are used to assign individuals mixed in the feeding grounds or along migratory corridors to their natal rookeries (Giovannotti et al. Citation2010; Clusa et al. Citation2014; Karaa et al. Citation2016), using mixed stock analysis (MSA; Grant et al. Citation1980).
Based on the results obtained from an integration of the different approaches mentioned above, Wallace et al. (Citation2011) proposed the definition of several regional management units (RMUs) to develop more specific and suitable strategies for the management and conservation of threatened globally distributed species, such as sea turtles. At the global scale, 10 RMUs were identified for C. caretta (Wallace et al. Citation2011), some of them also divisible into different rookery clusters or management units (MUs; Shamblin et al. Citation2014). Specifically, the Mediterranean RMU is not represented by a genetically homogeneous population, but consists of at least seven distinct MUs (Shamblin et al. Citation2014).
Within the Mediterranean Sea, nesting areas are mainly located in the eastern part of the basin, whereas the shallow waters (< 100 m) of the continental shelf, with their rich benthic communities (North Adriatic Sea, Gulf of Gabés, South Turkey and Egypt), are considered the key feeding habitats in the Mediterranean (Margaritoulis et al. Citation2003; Casale & Margaritoulis Citation2010). Usually, individuals from distinct Mediterranean and/or Atlantic rookeries are admixed in these feeding grounds (Laurent et al. Citation1998; Carreras et al. Citation2006; Clusa et al. Citation2014), except for the Adriatic foraging area in which only loggerhead turtles coming from Mediterranean rookeries have been found so far (Giovannotti et al. Citation2010; Yilmaz et al. Citation2012; Clusa et al. Citation2014). Therefore, knowledge of the biology and migrations of C. caretta represents a crucial point to evaluate the impact of different fishing activities in different areas and to identify possible management measures (Lucchetti et al. Citation2016).
The genetic composition within some feeding grounds, such as the Adriatic ones, is not completely known. Indeed, Casale et al. (Citation2012) highlighted that the Gulf of Manfredonia, Southwestern Adriatic, can be considered an additional important neritic feeding ground for C. caretta. The Gulf of Manfredonia is a small area characterised by shallow waters and frequented by a high number of loggerhead turtles of different ages (Casale et al. Citation2012). The high fishing pressure in these waters reduces the survival probability of sea turtles in this area (Casale et al. Citation2015). A recent study that describes the movement patterns of C. caretta within the Gulf of Manfredonia was performed by Casale and Simone (Citation2017) using satellite tracking, but it was not focused on reproductive migratory routes of loggerhead turtles foraging in this feeding ground.
In order to characterise genetically the loggerhead turtles frequenting the Southwestern Adriatic Sea, we analysed an 815-bp fragment of the mtDNA control region (D-loop) of 239 individuals collected in the Gulf of Manfredonia. Specifically, genetic analyses and an MSA were performed to: (1) describe the haplotype composition in the Southwestern Adriatic feeding ground; (2) reveal the contribution of known rookeries to this forging area; and (3) hypothesise migration routes within the Mediterranean Basin.
Materials and methods
A total of 239 individuals of Caretta caretta (see Table S1) rescued at the Manfredonia “Sea Turtle Rescue Centre” (CRTM; Foggia, Italy), managed by Legambiente Onlus, were analysed. Specimens were bycaught during fishing activity by local fishermen, or recovered from the shore after stranding along the Southern Adriatic coast of Italy. For each specimen, curved carapace length notch-to-tip (CCLn-t; Bolten Citation1999), weight, sex (if possible), sampling date, locality and distance from the coast at which turtles were caught, were recorded (Table S1).
For genetic analyses, blood or tissue samples were collected and preserved following the protocol proposed by Seutin et al. (Citation1991), and the DNA extraction was performed using a standard phenol–chloroform protocol (Taggart et al. Citation1992). After DNA extraction, polymerase chain reaction (PCR) was used to amplify an 815-bp fragment of the mtDNA (D-loop) using primers designed by Abreu-Grobois et al. (Citation2006): LCM15382 (5ʹ – GCT TAA CCC TAA AGC ATT GG – 3ʹ) and H950 (5ʹ – GTC TCG GAT TTA GGG GTT T – 3ʹ). The amplification was performed for each sample in a 25-μL PCR reaction containing: 1X MyTaq (Bioline) reaction buffer, 0.4 μM of each primer, 1.5 U MyTaqTM DNA polymerase (Bioline) and 3 μL of DNA (~40 ng/μL). Cycling conditions required an initial denaturation at 94°C for 5 min, followed by 35 cycles of 94°C for 60 s, 55°C for 60 s and 72°C for 60 s, with a final extension step at 72°C for 5 min. All PCR products were visualised on a 2% agarose gel stained with Gel RedTM (Biotium, Inc.) to evaluate amplification success, and digested at 37°C overnight using the restriction enzyme AluI in a 20-μL reaction containing 1X Tango Buffer, 7 U AluI and 2 μL of PCR product. The restriction enzyme AluI was chosen on the basis of the result obtained from a virtual digestion using the Sequence Manipulation Suite v. 2 (Stothard Citation2000) carried out on known D-loop haplotypes sequences. This simulation demonstrated the presence of two restriction sites that produce three different fragments (443, 360 and 54 bp in length). The following step was a single-strand conformation polymorphism (SSCP) analysis, which required the run of the digested products on a non-denaturing 8% polyacrylamide gel and successive visualisation by silver staining protocol (Benbouza et al. Citation2006). One sample (or, when possible, three) for each SSCP haplotype obtained was purified with the ExoSAP-it KIT and Sanger sequenced with both forward and reverse primers on an ABIPRISM 3730XL DNA sequencer (Applied Biosystem). All obtained D-loop sequences were aligned using the software ClustalW v. 1.8 (Thompson et al. Citation1994) with haplotypes previously described for this species and gathered by the Archie Carr Center for Sea Turtle Research of the University of Florida (ACCSTR; http://accstr.ufl.edu/files/cclongmtdna.pdf ). Novel haplotypes identified in this study were submitted to the ACCSTR for international nomenclature assignment and their nucleotide sequences were deposited in GenBank. The haplotype alignment file was imported into the software MEGA v. 7 (Kumar et al. Citation2016) to generate a phylogenetic tree. The best substitution model was selected on the basis of the lowest Bayesian information criterion (BIC) value and the phylogenetic analysis was performed using the neighbour-joining (NJ) method with the selected substitution model, Tamura 3-parameter with Gamma distribution (T92 + G), and 1000 bootstrap replicates to obtain statistical support values for the nodes. Only bootstrap values ≥ 50% were considered statistically significant (Margush & McMorris Citation1981). Finally, the proportion of individuals in the feeding ground contributed by different nesting areas was assessed using MSA, which was carried out using the rookery haplotype data set (Table S2) from Shamblin et al. (Citation2014). This analysis was performed using the Mixstock package v. 0.9.5.1 (Bolker Citation2012) on R v. 2.14.1 (http://www.r-project.org/). The Gelman–Rubin shrink factor (Gelman et al. Citation1996) was applied to test the algorithm convergence that was considered reached at 100,000 iterations, when a < 1.2 shrink factor value was obtained.
Results
Most of the Caretta caretta specimens recovered by the CRTM of Manfredonia and analysed in this study were caught as bycatch of bottom trawling activities (87.03%), while the remainder were collected from stranding events (8.79%), midwater trawling (3.35%) and gillnetting (0.42%). For 0.42% no information about the mode of recovery was available (Table S1). Observations made on three individuals (1.25% of the total) stranded dead on the Vieste beach in October 2013 allowed their classification as “hatchlings” on the basis of their carapace dimension (measured only as total curved carapace length, TCCL = 4.7 cm; Table S1) and by the presence of yolk sac residues. For the remaining C. caretta samples here analysed, morphometric analysis allowed the estimation of CCLn-t, which ranged from 10.5 to 88 cm (Table S1) and allowed the subdivision of these specimens into three arbitrary dimensional classes: (1) CCLn-t < 40 cm (small juveniles), including 19.67% of analysed specimens; (2) 40 cm ≤ CCLn-t ≤ 70 cm (large juveniles), including 66.11% of specimens (Luschi & Casale Citation2014); and (3) CCLn-t > 70 cm (mature adults; Margaritoulis et al. Citation2003; Casale et al. Citation2005), including 12.13% of specimens; while for two individuals (0.84%) no measurements were carried out (Table S1). Moreover, the young age of several individuals (51.88% of the sample) and the lack of necropsy did not allow their sex identification. Overall, 18.41% of the sample was composed of individuals identified as males and 29.71% as females (Table S1).
After genetic analyses of C. caretta individuals, a total of nine different SSCP haplotypes, confirmed by DNA sequencing, were detected. Seven of them were previously described in other studies (e.g. Shamblin et al. Citation2014) and two were observed here for the first time: CC-A71.1 and CC-A2.12 (GenBank Accession nos. KX929314 and KX929315, respectively). Among the haplotypes already described, CC-A2.1 was the most common (87.87% of the sample), followed by CC-A3.1 (4.18%), CC-A2.9 (2.93%), CC-A6.1 (2.93%), CC-A32.1 (0.42%), CC-A2.8 (0.42%) and CC-A10.1 (0.42%). Moreover, 0.42% of the loggerhead turtles analysed were assigned to the new haplotype CC-A71.1, and 0.42% to CC-A2.12. The phylogenetic analysis performed on C. caretta samples with the NJ method indicated the presence of two different mitochondrial lineages (called I and II; ), also described in Shamblin et al. (Citation2014). The two new haplotypes (CC-A71.1 and CC-A2.12) belonged to lineage II and, as shown by branch length, appeared to be phylogenetically close to haplotypes CC-A26.1 and CC-A2.1, respectively (). Interestingly, from a geographical point of view, haplotype CC-A26.1 was already observed in some rookeries along the Libyan coast, whereas haplotype CC-A2.1 was found to be widely distributed in Atlantic and Mediterranean rookeries (e.g. Shamblin et al. Citation2014).
Figure 1. Neighbour-joining phylogenetic tree that highlights the two D-loop haplogroups also described in Shamblin et al. (Citation2014), and the position of the new haplotypes (▲ and ●) identified in this study for the first time.

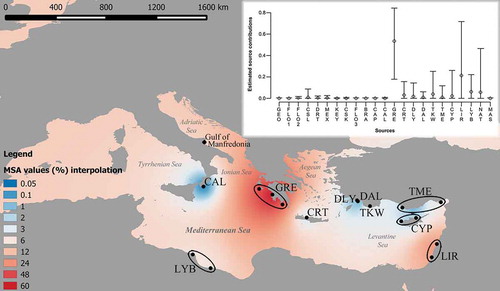
MSA results highlighted that the feeding grounds located along the Southern Italy Adriatic coast (mainly the Gulf of Manfredonia) host sea turtles likely coming from different rookeries located along the Mediterranean and Atlantic coasts (). In detail, the vast majority of the samples analysed come from Greek rookeries (53.58%), followed by Middle-East Mediterranean (21.11%) and Libyan rookeries (5.94%). Finally, less than 1% of the individuals analysed showed the CC-A10.1 haplotype, so far detected in some Atlantic rookeries of the Gulf of Mexico and Florida (Shamblin et al. Citation2012).
Figure 2. The inset graph shows the estimated contribution of the globally described rookeries (Shamblin et al. Citation2014) to the Southwestern Adriatic feeding ground. Bars represent 95% confidence intervals. The map shows with different colours the contribution (as a percentage) of the Mediterranean rookeries. GEO (Cape Island, South Carolina + Ossabaw Island, Georgia), FLO1 (Canaveral National Seashore + Melbourne Beach, Florida), FLO2 (Juno Beach + Ft. Lauderdale, Florida), CSL (Cay Sal, Bahamas), DRT (Dry Tortugas, Florida), MEX (Isla Cozumel + Quintana Roo mainland, Mexico), KEY (Keewaydin Island, Florida), CSK (Casey Key, Florida), FLO3 (St. George Island + Cape San Blas, Florida), BRA (Sergipe + Bahia + Espírito Santo + Rio de Janeiro, Brazil), CAP (Boa Vista + Sal + Santa Luzia + Maio, Cape Verde), CAL (Calabria, Italy), GRE (Zakynthos Island + Kyparissia + Lakonikos, Greece), CRT (Rethymno, Crete), DLY (Dalyan, Turkey), DAL (Dalaman, Turkey), TKW (western Turkey), TME (middle Turkey + eastern Turkey), CYP (Alagadi + Akamas, Cyprus), LIR (El Mansouri, Lebanon + Israel), LYB (Sirte + Misurata, Lybia), NAT (Tongaland, KwaZulu-Natal, South Africa), MAS (Masirah, Oman).
Discussion
In this study, the haplotype composition of the Caretta caretta sample from the Southwestern Adriatic feeding ground and the haplotype composition of the main rookeries known around the world (Shamblin et al. Citation2014) were compared by MSA. This test highlighted that loggerhead turtles sampled in the above feeding ground principally come from Greek rookeries (53.58%), as already demonstrated in other studies (Giovannotti et al. Citation2010; Yilmaz et al. Citation2012; Garofalo et al. Citation2013), confirming the existence of a short-range migratory route that C. caretta follows to enter the Adriatic Sea (Giovannotti et al. Citation2010). A secondary contribution to the Adriatic feeding ground is provided by the Lebanon and Israel rookeries (21.11%). This result contrasts with the conclusions by Giovannotti et al. (Citation2010) who indicated the Turkish rookeries were the second most important contributors. This inconsistency could be due to the use, by Giovannotti et al. (Citation2010), of a shorter D-loop sequence (~380 bp) that does not allow one to distinguish the CC-A2.9 haplotype, common in Israel, Lebanon and Libyan rookeries, from the cosmopolitan CC-A2.1 haplotype. On the contrary, in this study the CC-A2.9 haplotype was observed in seven individuals.
Moreover, the MSA also highlighted another possible migration route from Libya to the Adriatic coasts (5.94%), confirmed by the recovery of the CC-A2.9 haplotype which is common in the Libyan nesting sites, and by the recovery of the new CC-A71.1 haplotype (see below). A contribution from the rookeries of the Gulf of Sirte to the Adriatic feeding ground was confirmed by other previous studies (Giovannotti et al. Citation2010; Garofalo et al. Citation2013). Interestingly, the difference in the estimated source contributions between Middle-East and Libyan rookeries (respectively, 21.11 vs 5.94%) seems to be too high because the CC-A2.9 haplotype is more common in Libya. However, it should be noted that both values were characterised by wide confidence limits (). Therefore, the interpretation of the MSA in these circumstances should be made with great caution (e.g. Rees et al. Citation2017). Similarly, a wide confidence limit characterised also the unexpected source contribution from the South African rookery (NAT, 5.51%; ). This geographic anomaly was probably due to the presence of the sole common haplotype CC-A2.1 observed in NAT (Shamblin et al. Citation2014). According to Jensen et al. (Citation2013) the presence of common mtDNA haplotypes shared among rookeries could bias the results of MSA. For this reason, the role of South African rookeries was not further commented on.
Generally, MSA has allowed the identification of the Adriatic Basin as a feeding ground mainly used by C. caretta individuals coming from the mid-eastern Mediterranean Basin. However, the recovery in this study of the CC-A10.1 haplotype is very interesting because this is common only along the coasts of the Gulf of Mexico and Florida (Shamblin et al. Citation2012) and represents the first observation of Atlantic loggerhead turtles in the Adriatic feeding ground, whereas the previous observations of Atlantic individuals into the Mediterranean Sea were made in the Tyrrhenian Sea and Lampedusa Island (Casale et al. Citation2002; Garofalo et al. Citation2013). This finding highlights the importance of the Adriatic Basin as a foraging area, for C. caretta not only from the mid-eastern Mediterranean rookeries, but also from Atlantic rookeries. Haplotypes CC-A2.12 and CC-A71.1 were described for the first time in this study, and for this reason it was not possible to determine which rookeries they are from. However, a Libyan origin can be hypothesised for CC-A71.1 based on its phylogenetic affinity with CC-A26.1 that was observed only along the Libyan coast (Shamblin et al. Citation2014), whereas it is not easy to formulate a hypothesis for the origin of CC-A2.12 because it is closely related to the cosmopolitan CC-A2.1 haplotype.
Finally, relevant information may also be obtained from the morphological analyses of sampled individuals. Firstly, the observation of individuals belonging to typical size classes of large juveniles and adults, and the recovery mode principally as bottom trawling bycatch, have confirmed, according to Casale et al. (Citation2012), the importance of the Gulf of Manfredonia mainly as a neritic foraging ground and growing area. Secondly, the recovery on the Vieste beach of three stranded hatchlings, with reduced carapace length (TCCL = 4.7 cm) and evident residues of the yolk sac, suggests that the Southern Italy Adriatic coasts, despite the high human impact, could host nesting areas of the loggerhead turtles. This observation is confirmed by the identification along the Apulian coasts of numerous nesting sites (Marra et al. Citation2015; Garofalo et al. Citation2016) and by the description of 13 hatchlings in October 2013 stranded on the Vieste beach (Marra et al. Citation2015), to which the three dead hatchlings analysed here seem to belong. In addition, according to a recent study based on satellite tracking of five loggerhead turtles caught in the Gulf of Manfredonia (Casale & Simone Citation2017), the hypothesis of the presence in this area of a new permanent nesting site appears plausible. However, further studies will be indispensable to fully understand whether this area is only a sporadic or a new permanent nesting site, or whether it was determined by a recent expansion of the nesting areas due to climate changes (Witt et al. Citation2010; Pikesley et al. Citation2015). Furthermore, genetic analyses highlighted that all the hatchlings bear the CC-A2.1 haplotype, the most common in the Atlantic and Mediterranean rookeries. This result does not allow the formulation of any hypotheses regarding the genetic relationship between the loggerhead turtles nesting along the Apulian coast and those nesting in different areas.
In any case, this study proves the growing importance of the Southwestern Adriatic Sea as a nesting area. Moreover, in countertendency with the general decline of sea turtles at the global scale, the number of turtles rescued at the Adriatic CRTMs has increased over the last five years (Marine Environment Department Database, Italian Ministry of Environment). These results are a matter of great concern for the conservation of C. caretta, because they suggest that the Adriatic Sea is becoming a “funnel” for turtles by attracting them towards a closed basin characterised by high fishing pressure (Lucchetti et al. Citation2016). Therefore, the high density of turtles in a small area with high fishing activity makes the impact of bycatch by fishing particularly intense on this species.
Concluding remarks
The Eastern part of the Mediterranean Sea is a well-known nesting area for Caretta caretta, whereas the Adriatic Sea is generally considered an important feeding ground for this species. In this research, results obtained from the analysis of D-loop sequences from 239 individuals of C. caretta allowed us to verify that the feeding grounds of the Southwestern Adriatic basin host loggerhead turtles coming from different rookeries that are located mainly in Greece and, to a lesser extent, in Lebanon, Israel and Libya, and also on Atlantic coasts. These results provide more detailed information about the routes followed during feeding migrations. Unfortunately, it was not possible to link, through the use of the MSA, the two “orphan haplotypes” to specific nesting areas due to the lack of knowledge about the existing rookeries. This weakness highlights the need to improve the genetic characterisation of nesting areas, in order to better identify migratory routes and to develop reliable conservation strategies for this species. In addition, the recovery of three hatchlings along the Vieste beach, an unusual event at these latitudes, highlights the growing importance of the Adriatic area not only as a feeding ground for C. caretta, but also as a new nesting site. Finally, the identification of which natal rookeries contribute to the Southern Italy Adriatic feeding ground is important to understand which nesting sites are mostly impacted by the human activities carried out in this area and/or along the putative migratory corridors. This information, together with the knowledge of the presence of new probable nesting sites along the Southwestern Adriatic coast, is important for planning future programmes for the monitoring and conservation of this species, and for developing new management measures and mitigation strategies to reduce bycatch in fishing operations in order to protect the most impacted rookeries and feeding grounds within the Mediterranean Sea.
Acknowledgements
All authorisations to handle and analyse samples were provided by the Italian “Ministero dell’Ambiente e della Tutela del Territorio e del Mare”, 0040625-26/11/2012-PNM-II and 0037650-31/05/2013/PNM-II. This research was supported by funds (Ricerca Scientifica d’Ateneo 2014) provided by Università Politecnica delle Marche (Ancona, Italy) to Prof. Vincenzo Caputo Barucchi and Dr Massimo Giovannotti.
Supplemental data
Supplemental data for this article can be accessed here.
References
- Abreu-Grobois FA , Horrocks J , Formia A , Dutton PH , LeRoux R , Vélez-Zuazo X , Soares L , Meylan P 2006. New mtDNA Dloop primers which work for a variety of marine turtle species may increase the resolution of mixed stock analyses. Proceedings of the 26th Annual Symposium on Sea Turtle Biology Conservation, 3–8 April 2006, Crete. p. 179.
- Benbouza H , Jacquemin JM , Baudoin JP , Mergeai G. 2006. Optimization of reliable, fast, cheap and sensitive silver staining method to detect SSR markers in polyacrylamide gels. Biotechnology, Agronomy, Society and Environment 10:77–81.
- Bentivegna F. 2002. Intra-Mediterranean migrations of loggerhead sea turtles (Caretta caretta) monitored by satellite telemetry. Marine Biology 141:795–800. DOI: 10.1007/s00227-002-0856-z.
- Bolker B 2012. mixstock: functions for mixed stock analysis. R package version 0.9.5.1. Available: http://cran.r-project.org/web/packages/mixstock/index.html . Accessed Oct 2017 30.
- Bolten AB . 1999. Techniques for measuring sea turtles. In: Eckert KL , Bjorndal KA , Abreu-Grobois FA , Donnelly M , editors. Research and management techniques for the conservation of sea turtles. Washington: IUCN/SSC Marine Turtle Specialist Group. pp. 110–114.
- Bolten AB . 2003. Variation in sea turtle life history patterns: Neritic versus oceanic developmental stages. In: Lutz PL , Musick JA , Wyneken J , editors. The biology of sea turtles. Boca Raton: CRC Press. pp. 243–257.
- Bowen BW , Kamezaki N , Limpus CJ , Hughes GR , Meylan AB , Avise JC . 1994. Global phylogeography of the loggerhead turtle (Caretta caretta) as indicated by mitochondrial DNA haplotypes. Evolution 48:1820–1828.
- Broderick AC , Glen F , Godley BJ , Hays GC . 2002. Estimating the number of green and loggerhead turtles nesting annually in the Mediterranean. Oryx 36:227–235. DOI: 10.1017/S0030605302000431.
- Carreras C , Pascual M , Cardona L , Aguilar A , Margaritoulis D , Rees A , Turkozan O , Levy Y , Gasith A , Aureggi M , Khalil M . 2007. The genetic structure of the loggerhead sea turtle (Caretta caretta) in the Mediterranean as revealed by nuclear and mitochondrial DNA and its conservation implications. Conservation Genetics 8:761–775. DOI: 10.1007/s10592-006-9224-8.
- Carreras C , Pont S , Maffucci F , Pascual M , Barceló A , Bentivegna F , Cardona L , Alegre F , SanFélix M , Fernández G , Aguilar A . 2006. Genetic structuring of immature loggerhead sea turtles (Caretta caretta) in the Mediterranean Sea reflects water circulation patterns. Marine Biology 149:1269–1279. DOI: 10.1007/s00227-006-0282-8.
- Casale P . 2011. Sea turtle by-catch in the Mediterranean. Fish and Fisheries 12:299–316. DOI: 10.1111/j.1467-2979.2010.00394.x.
- Casale P , Freggi D , Basso R , Argano R . 2005. Size at male maturity, sexing methods and adult sex ratio in loggerhead turtles (Caretta caretta) from Italian waters investigated through tail measurements. Journal of Herpetology 15:145–148.
- Casale P , Freggi D , Basso R , Vallini C , Argano R . 2007. A model of area fidelity, nomadism, and distribution patterns of loggerhead sea turtles (Caretta caretta) in the Mediterranean Sea. Marine Biology 152:1039–1049. DOI: 10.1007/s00227-007-0752-7.
- Casale P , Freggi D , Cinà A , Rocco M . 2013. Spatio-temporal distribution and migration of adult male loggerhead sea turtles (Caretta caretta) in the Mediterranean Sea: Further evidence of the importance of neritic habitats off North Africa. Marine Biology 160:703–718. DOI: 10.1007/s00227-012-2125-0.
- Casale P , Freggi D , Furii G , Vallini C , Salvemini P , DeFlorio M , Totaro G , Raimondi S , Fortuna C , Godley B . 2015. Annual survival probabilities of juvenile loggerhead sea turtles indicate high anthropogenic impact on Mediterranean populations. Aquatic Conservation: Marine and Freshwater Ecosystems 25:690–700. DOI: 10.1002/aqc.2467.
- Casale P , Laurent L , Gerosa G , Argano R . 2002. Molecular evidence of male-biased dispersal in loggerhead turtle juveniles. Journal of Experimental Marine Biology and Ecology 267:139–145. DOI: 10.1016/S0022-0981(01)00365-3.
- Casale P , Margaritoulis D . 2010. Sea turtles in the Mediterranean: distribution, threats and conservation priorities. Gland, Switzerland: IUCN/SSC Marine Turtle Specialist Group.
- Casale P , Simone G . 2017. Seasonal residency of loggerhead turtles Caretta caretta tracked from the Gulf of Manfredonia, South Adriatic. Mediterranean Marine Science 18:4–10. DOI: 10.12681/mms.1663.
- Casale P , Simone G , Conoscitore C , Conoscitore M , Salvemini P . 2012. The Gulf of Manfredonia: A new neritic foraging area for loggerhead sea turtles in the Adriatic Sea. Acta Herpetologica 7:1–12.
- Claudet J , Fraschetti S . 2010. Human-driven impacts on marine habitats: A regional meta-analysis in the Mediterranean Sea. Biological Conservation 143:2195–2206. DOI: 10.1016/j.biocon.2010.06.004.
- Clusa M , Carreras C , Pascual M , Demetropoulos A , Margaritoulis D , Rees AF , Hamza AA , Khalil M , Aureggi M , Levy Y , Türkozan O , Marco A , Aguilar A , Cardona L . 2013. Mitochondrial DNA reveals Pleistocenic colonisation of the Mediterranean by loggerhead turtles (Caretta caretta). Journal of Experimental Marine Biology and Ecology 439:15–24. DOI: 10.1016/j.jembe.2012.10.011.
- Clusa M , Carreras C , Pascual M , Gaughran SJ , Piovano S , Giacoma C , Fernández G , Levy Y , Tomás J , Raga JA , Maffucci F , Hochscheid S , Aguilar A , Cardona L . 2014. Fine-scale distribution of juvenile Atlantic and Mediterranean loggerhead turtles (Caretta caretta) in the Mediterranean Sea. Marine Biology 161:509–519. DOI: 10.1007/s00227-013-2353-y.
- Coll M , Piroddi C , Albouy C , Ben Rais Lasram F , Cheung WWL , Christensen V , Karpouzi VS , Guilhaumon F , Mouillot D , Paleczny M , Palomares ML , Steenbeek J , Trujillo P , Watson R , Pauly D . 2012. The Mediterranean Sea under siege: Spatial overlap between marine biodiversity, cumulative threats and marine reserves. Global Ecology and Biogeography 21:465–480. DOI: 10.1111/j.1466-8238.2011.00697.x.
- Coll M , Piroddi C , Steenbeek J , Kaschner K , Ben Rais Lasram F , Aguzzi J , Ballesteros E , Bianchi CN , Corbera J , Dailianis T , Danovaro R , Estrada M , Froglia C , Galil BS , Gasol JM , Gertwagen R , Gil J , Guilhaumon F , Kesner-Reyes K , Kitsos MS , Koukouras A , Lampadariou N , Laxamana E , de la Cuadra CMLF , Lotze HK , Martin D , Mouillot D , Oro D , Raicevich S , Rius-Barile J , Saiz-Salinas JI , San Vicente C , Somot S , Templado J , Turon X , Vafidis D , Villanueva R , Voultsiadou E . 2010. The biodiversity of the Mediterranean Sea: Estimates, patterns and threats. PLoS ONE 5:e11842.
- Cuttelod A , Garcia N , Malak DA , Temple H , Katariya V . 2008. The Mediterranean: A biodiversity hotspot under threat. In: Hilton-Taylor and Stuart SN, editors. The 2008 Review of the IUCN Red List of Threatened Species. Gland, Switzerland.
- Encalada S , Bjorndal K , Bolten A , Zurita JC , Schroeder B , Possardt E , Sears CJ , Bowen BW . 1998. Population structure of loggerhead turtle (Caretta caretta) nesting colonies in the Atlantic and Mediterranean as inferred from mitochondrial DNA control region sequences. Marine Biology 130:567–575. DOI: 10.1007/s002270050278.
- Garofalo L , Marzano G , Caputo A , Carlino P , Olivieri V , Lorenzini R 2016. Locations of summer meetings…for sea turtles too! Mitochondrial characterization of Caretta caretta nests from the beaches of the southern-middle Adriatic and the northern Ionian Seas. Book of abstracts XI Congresso Nazionale della Societas Herpetologica Italica, 22–25 Settembre 2016, Trento. pp. 131–132.
- Garofalo L , Mastrogiacomo A , Casale P , Carlini R , Eleni C , Freggi D , Gelli D , Knittweis L , Mifsud C , Mingozzi T , Novarini N , Scaravelli D , Scillitani G , Oliverio M , Novelletto A . 2013. Genetic characterization of central Mediterranean stocks of the loggerhead turtle (Caretta caretta) using mitochondrial and nuclear markers, and conservation implications. Aquatic Conservation: Marine and Freshwater Ecosystems 23:868–884. DOI: 10.1002/aqc.v23.6.
- Gelman A , Carlin J , Stern HS , Rubin DB . 1996. Bayesian data analysis. New York, NY: Chapman and Hall.
- Giovannotti M , Franzellitti S , Nisi Cerioni P , Fabbri E , Guccione S , Vallini C , Tinti F , Caputo V . 2010. Genetic characterization of loggerhead turtle (Caretta caretta) individuals stranded and caught as bycatch from the North-Central Adriatic Sea. Amphibia-Reptilia 31:127–133. DOI: 10.1163/156853810790457902.
- Girard C , Tucker AD , Calmettes B . 2009. Post-nesting migrations of loggerhead sea turtles in the Gulf of Mexico: Dispersal in highly dynamic conditions. Marine Biology 156:1827–1839. DOI: 10.1007/s00227-009-1216-z.
- Grant WS , Milner GB , Krasnowski P , Utter FM . 1980. Use of biochemical genetic variants for identification of sockeye salmon (Oncorhynchus nerka) stocks in Cook Inlet, Alaska. Canadian Journal of Fisheries and Aquatic Sciences 37:1236–1247. DOI: 10.1139/f80-159.
- Hatase H , Kinoshita M , Bando T , Kamezaki N , Sato K , Matsuzawa Y , Goto K , Omuta K , Nakashima Y , Takeshita H , Sakamoto W . 2002. Population structure of loggerhead turtles, Caretta caretta, nesting in Japan: Bottlenecks on the Pacific population. Marine Biology 141:299–305. DOI: 10.1007/s00227-002-0819-4.
- Jensen MP , FitzSimmons NN , Dutton PH . 2013. Molecular genetics of sea turtles. In: Wyneken J , Lohmann KJ , Musick JA , editors. The biology of sea turtles, Volume 3. Boca Raton: CRC Press. pp. 135–154.
- Karaa S , Maffucci F , Jribi I , Bologna MA , Borra M , Biffali E , Bradai MN , Hochscheid S . 2016. Connectivity and stock composition of loggerhead turtles foraging on the North African continental shelf (Central Mediterranean): Implications for conservation and management. Marine Ecology 37:1103–1115. DOI: 10.1111/maec.2016.37.issue-5.
- Kumar S , Stecher G , Tamura K . 2016. MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Molecular Biology and Evolution 33:1870–1874. DOI: 10.1093/molbev/msw054.
- Laurent L , Casale P , Bradai MN , Godley BJ , Broderick AC , Schroth W , Schierwater B , Levy AM , Freggi D , Abd El-Mawla EM , Hadoud DA , Gomati HE , Domingo M , Hadliehristophorou M , Kornaraky L , Demirayak F , Gautier CH . 1998. Molecular resolution of marine turtle stock composition in fishery bycatch: A case study in the Mediterranean. Molecular Ecology 7:1529–1542. DOI: 10.1046/j.1365-294x.1998.00471.x.
- Lewison RL , Crowder LB , Read AJ , Freeman SA . 2004. Understanding impacts of fisheries bycatch on marine megafauna. Trends in Ecology & Evolution 19:598–604. DOI: 10.1016/j.tree.2004.09.004.
- Lewison RL , Crowder LB , Wallace BP , Moore JE , Cox T , Zydelis R , McDonald S , DiMatteo A , Dunn DC , Kott CY , Bjorkland R , Kelez S , Soykan C , Stewart KR , Sims M , Boustany A , Read AJ , Halpin P , Nichols WJ , Safina C 2014. Global patterns of marine mammal, seabird, and sea turtle bycatch reveal taxa-specific and cumulative megafauna hotspots. Proceedings of the National Academy of Sciences 111:5271–5276.
- Lucchetti A , Pulcinella J , Angelini V , Pari S , Russo T , Cataudella S . 2016. An interaction index to predict turtle bycatch in a Mediterranean bottom trawl fishery. Ecological Indicators 60:557–564. DOI: 10.1016/j.ecolind.2015.07.007.
- Luschi P , Casale P . 2014. Movement patterns of marine turtles in the Mediterranean Sea: A review. Italian Journal of Zoology 81:478–495. DOI: 10.1080/11250003.2014.963714.
- Marcovaldi MÂ , Lopez GG , Soares LS , Lima EHSM , Thom JCA , Almeida AP . 2010. Satellite-tracking of female loggerhead turtles highlights fidelity behavior in northeastern Brazil. Endangered Species Research 12:263–272. DOI: 10.3354/esr00308.
- Margaritoulis D , Argano R , Baran I , Bentivegna F , Bradai MN , Camiñas JA , Casale P , De Metrio G , Demetropoulos A , Gerosa G , Godley BJ , Haddoud DA , Houghton J , Laurent L , Lazar B . 2003. Loggerhead turtles in the Mediterranean Sea: Present knowledge and conservation perspectives. In: Bolten AB , Witherington BE , editors. Loggerhead sea turtle. Washington: Smithsonian Books. pp. 175–198.
- Margush T , McMorris FR . 1981. Consensus n-trees. Bullettin of Mathematical Biology 43:239–244.
- Marra M , Scillitani G , Flore G , Zaccaria G , Capodiferro T , Ciccolella A , Marzano G , De Franco F , Corriero G . 2015. Analisi cartografica preliminare delle nidificazioni (2006-2013) e realizzazione di un database degli spiaggiamenti (1994-2013) della Tartaruga comune Caretta caretta lungo le coste pugliesi. Atti X Congresso Nazionale Societas Herpetologica Italica, Genova. pp. 393–398.
- Moncada F , Abreu-Grobois FA , Bagley D , Bjorndal KA , Bolten AB , Camiñas JA , Ehrhart L , Muhlia-Melo A , Nodarse G , Schroeder BA , Zurita J , Hawkes LA . 2010. Movement patterns of loggerhead turtles Caretta caretta in Cuban waters inferred from flipper tag recaptures. Endangered Species Research 11:61–68. DOI: 10.3354/esr00248.
- Myers N , Mittermeier RA , Mittermeier CG , Da Fonseca GAB , Kent J . 2000. Biodiversity hotspots for conservation priorities. Nature 403:853–858. DOI: 10.1038/35002501.
- Pikesley SK , Broderick AC , Cejudo D , Coyne MS , Godfrey MH , Godley BJ , Lopez P , López-Jurado LF , Merino SE , Varo-Cruz N , Witt MJ , Hawkes LA . 2015. Modelling the niche for a marine vertebrate: A case study incorporating behavioural plasticity, proximate threats and climate change. Ecography 38:803–812. DOI: 10.1111/ecog.2015.v38.i8.
- Rees AF , Carreras C , Broderick AC , Margaritoulis D , Stringell TB , Godley BJ . 2017. Linking loggerhead locations: Using multiple methods to determine the origin of sea turtles in feeding grounds. Marine Biology 164:30. DOI: 10.1007/s00227-016-3055-z.
- Revelles M , Camiñas JA , Cardona L , Parga M , Tomás J , Aguilar A , Alegre F , Raga A , Bertolero A , Oliver G . 2008. Tagging reveals limited exchange of immature loggerhead sea turtles (Caretta caretta) between regions in the western Mediterranean. Scientia Marina 72:511–518.
- Seutin G , White BN , Boag PT . 1991. Preservation of avian blood and tissue samples for DNA analyses. Canadian Journal of Zoology 69:82–90. DOI: 10.1139/z91-013.
- Shamblin BM , Bolten AB , Abreu-Grobois FA , Bjorndal KA , Cardona L , Carreras C , Clusa M , Monzón-Argüello C , Nairn CJ , Nielsen JT , Nel R , Soares LS , Stewart KR , Vilaça ST , Türkozan O , Yilmaz C , Dutton PH . 2014. Geographic patterns of genetic variation in a broadly distributed marine vertebrate: New insights into loggerhead turtle stock structure from expanded mitochondrial DNA sequences. PLoS One 9:e85956. DOI: 10.1371/journal.pone.0085956.
- Shamblin BM , Bolten AB , Bjorndal KA , Dutton PH , Nielsen JT , Abreu-Grobois FA , Reich KJ , Witherington BE , Bagley DA , Ehrhart LM . 2012. Expanded mitochondrial control region sequences increase resolution of stock structure among North Atlantic loggerhead turtle rookeries. Marine Ecology Progress Series 469:145–160. DOI: 10.3354/meps09980.
- Stothard P . 2000. The sequence manipulation suite: JavaScript programs for analyzing and formatting protein and DNA sequences. Biotechniques 28:1102–1104.
- Taggart JB , Hynes RA , Prodouhl PA , Ferguson A . 1992. A simplified protocol for routine total DNA isolation from salmonid fishes. Journal of Fish Biology 40:963–965. DOI: 10.1111/jfb.1992.40.issue-6.
- Thompson JD , Higgins DG , Gibson TJ . 1994. CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Research 22:4673–4680. DOI: 10.1093/nar/22.22.4673.
- Wallace BP , DiMatteo AD , Bolten AB , Chaloupka MY , Hutchinson BJ , Abreu-Grobois A , Mortimer JA , Seminoff JA , Amorocho D , Bjorndal KA , Bourjea J , Bowen BW , Dueñas RB , Casale P , Choudhury BC , Costa A , Dutton PH , Fallabrino A , Finkbeiner EM , Girard A , Girondot M , Hamann M , Hurley BJ , López-Mendilaharsu M , Marcovaldi MA , Musick JA , Nel R , PIlcher NJ , Troëng S , Witherington B , Mast RB . 2011. Global conservation priorities for marine turtles. PLoS ONE 6:e24510. DOI: 10.1371/journal.pone.0024510.
- Witt MJ , Hawkes LA , Godfrey MH , Godley BJ , Broderick AC . 2010. Predicting the impacts of climate change on a globally distributed species: The case of the loggerhead turtle. Journal of Experimental Biology 213:901–911. DOI: 10.1242/jeb.038133.
- Yilmaz C , Turkozan O , Bardakci F , White M , Kararaj E . 2012. Loggerhead turtles (Caretta caretta) foraging at Drini Bay in Northern Albania: Genetic characterization reveals new haplotypes. Acta Herpetologica 7:155–162.
