Abstract
Evidence available for most inland water and terrestrial organisms highlights the significant role played by southern Italy, Sicily and the Maltese islands as refuges during Pleistocene climatic fluctuations. However, to date, the hypothesis that these areas may have acted as Pleistocene refugia for the freshwater crab Potamon fluviatile has not been explicitly tested, and a recent origin of local P. fluviatile populations was proposed on the basis of a small set of analysed molecular data. We have thus expanded the currently available data set on the population genetic structure of P. fluviatile through dedicated samplings in Sicily (Italy, 18 specimens), the Maltese Islands (Malta, 15 specimens) and the island of Corfu (Greece, seven specimens), with the explicit aim of testing the role of southern Italy, Sicily and the Maltese archipelago as refugia where early Pleistocene or pre-Quaternary populations of the freshwater crab could have persisted to date. Based on the analysis of both novel and published cytochrome c oxidase subunit I (COI) sequence data, we gathered evidence supporting a recent origin of the southern Italian, Sicilian and Maltese populations of Potamon fluviatile from the Balkans, thus rejecting the hypothesised Pleistocene persistence in the area of pre-existing local populations of the same species.
Introduction
The freshwater crab family Potamidae is classified into two subfamilies: Potaminae (distributed across Europe, North Africa, Yemen, the Middle East and India) and Potamiscinae (Eastern and South-eastearn Asia) (Yeo & Ng Citation2003). A revision of the genus Potamon Savigny, 1816 in the West-Palearctic region lists 16 species, divided into four mostly allopatric or parapatric subgenera that were distinguished through morphological characters, mostly related to the gonopods (Brandis et al. Citation2000). In particular, the subgenus Euthelphusa Pretzmann, 1962, occurring in the western Mediterranean area and on the Balkan peninsula, includes the species Potamon fluviatile (Herbst, Citation1785), P. pelops Jesse et al., Citation2010 and P. algeriense Bott, Citation1967 (Brandis et al. Citation2000). P. fluviatile is distributed across Italy, Malta, Croatia, southern Dalmatia, Albania and Greece (Brandis et al. Citation2000), usually segregated from other autochthonous macro-decapods (Barbaresi et al. Citation2007; Mazza et al. Citation2017). Adults may reach a carapace length of about 50 mm over a 10-year-long lifespan (Gherardi et al. Citation1987; Micheli et al. Citation1990; Scalici et al. Citation2013); they are omnivorous, feeding primarily on invertebrates, amphibians and macrophytes (Cumberlidge Citation2008). Since 2008, P. fluviatile has been assessed as “Near threatened” on the International Union for Conservation of Nature (IUCN) Red List of Threatened Species ( http://www.iucnredlist.org/details/134293/0 ), because the populations of the species historically declined as a result of pollution, over-harvesting and mismanagement of habitats (Matthew & Reynolds Citation1995; Gherardi & Holdich Citation1999).
In the last decade, some studies were conducted on P. fluviatile in order to investigate its phylogenetic relationships with the other species within the subgenus Euthelphusa, and to elucidate its phylogeography (see Jesse et al. Citation2010 and references therein). Based on the available molecular data, Jesse et al. (2009, 2010, 2011) proposed a non-human-mediated late Pleistocene origin from the Balkans of the Italian and Maltese populations of the species. However, it is well known that, during Pleistocene glaciations, southern Italy, Sicily and the Maltese archipelago acted as refugia for different species which persisted in situ (Hewitt Citation2000, Citation2004), with a resulting genetic differentiation of these “southern” strains, for both aquatic (e.g. Canestrelli et al. Citation2007; Canestrelli & Nascetti Citation2008; Kindler et al. Citation2013; Vamberger et al. Citation2015) and terrestrial (e.g. Michaux et al. Citation2003; Bezerra et al. Citation2015) taxa.
The purpose of our study was thus to explicitly test the possible role played by southern Italy and Malta as refugia for P. fluviatile during the Pleistocene, through the implementation of an expanded sampling protocol of the genetic diversity of different populations of the species in Sicily and in the Maltese archipelago. If this geographic area did indeed act as a Pleistocene refugium for the species, we expect to find the presence of endemic haplotypes, possibly mixed with widespread ones shared with those of the Balkan populations of the species, as in the case of incomplete lineage sorting or as in the case of a secondary contact between the populations inhabiting the “southern Italian” and “Balkan” refugia during the late Pleistocene.
Material and methods
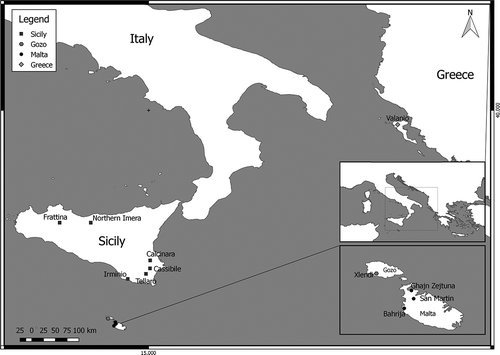
In total, 40 novel specimens (1–7 per site; see ) were analysed, taking one leg from each individual, and releasing them back in the same sampling site where they were collected. The loss of one leg does not constitute harm to the sampled individuals, since they are able to routinely regenerate any lost appendages after the next molt (cf. Das Citation2015 and references therein). Sampled legs were fixed in situ in 90% ethanol. In addition, 21 haplotype sequences, corresponding to 104 individuals collected throughout the known distribution range of the species, were downloaded from GenBank (accession numbers: EU908226–EU908246) and analysed along with our sequences. Further information on the sampling sites and number of sequenced individuals is given in . The map of the sampling sites (see ) was plotted using the QGIS software v. 2.18.2 ( http://www.qgis.org ).
Table I. Location of the sampling sites.
Genomic DNA was extracted from the muscle tissue of each leg using the Genomic DNA Extraction Kit (RBC BioScience) following the manufacturer’s protocol. The selective amplification of a 502-bp-long fragment from the gene encoding for the cytochrome oxidase subunit I (COI) was carried out by the polymerase chain reaction (PCR). The PCR mix consisted of 18.05 µL double-distilled water, 2.5 µL Buffer 10X including 15 mM MgCl2 solution, 0.25 µL dNTPs (10 mM of each), 0.9 µL of each primer (10 µM), 0.4 µL Taq Polymerase 5 u/µL and 2 µL of DNA template, for a total volume of 25 µL. The thermal cycle (35 cycles; denaturing 95°C for 45 s/annealing 46°C for 45 s/extension 72°C for 50 s) was conducted with the primers COL6b (5ʹ-ACA AAT CAT AAA GAT ATY GG-3ʹ) and COH6 (5ʹ-TAD ACT TCD GGR TGD CCA AAR AAY CA-3ʹ), described by Schubart and Huber (Citation2006). After PCR, 5 µL of each PCR product were separated by electrophoresis on a 2% agarose gel at 90 V for 20 minutes and visualised with a UV Transilluminator. When PCR products showed a clear and single band of the correct expected length, they were purified using the Exo-SAP-IT® kit (Affymetrix USB) and sequenced by Macrogen Inc. (Seoul, South Korea) with ABI 3130xL (Applied Biosystems) sequencer. The forward primers were used for direct sequencing of the PCR product. The quality of the obtained chromatograms was checked through the measurement of their Phred scores (Richterich Citation1998). Only sequences with continuous reads of high-quality bases Quality Value scores ((QV) > 20) were used. Sequences were analysed and manually proofread with the DNA sequencing software Chromas v. 2.6.2 (Technelysium, Pty. Ltd. 1998, Queensland, Australia) and aligned with ClustalX v. 2.1 (Larkin et al. Citation2007). MEGA v. 7.0 (Kumar et al. Citation2016) was used to translate the COI sequences into amino acids in order to check for the possible presence of frameshifts or stop codons, which would indicate the presence of sequencing errors or pseudogenes.
Upon aligning our novel sequences with those downloaded from GenBank, we had to trim out 35 bp from the sequences downloaded from GenBank, which were originally 537 bp long. This trimming led to the merging of some haplotypes (see Table S1 for correspondence of our haplotype codes with those described by Jesse et al. Citation2009).
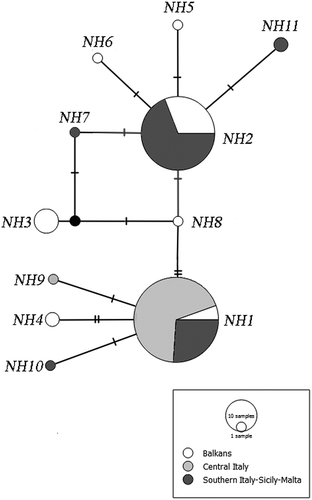
A haplotype network for the identified sequences was constructed through the software Popart (v. 1.7, http://popart.otago.ac.nz ), using the median-joining method (Bandelt et al. Citation1999) ().
Figure 2. Median-joining haplotype network based on a 502-bp-long fragment of the mitochondrial DNA gene Cytochrome oxidase subunit 1 (COI). Dashes indicate substitution steps. Each circle represents a haplotype and its size is proportional to its frequency.
We partitioned a priori the sampling sites into three geographical groups: (i) Balkans; (ii) Central Italy; and (iii) southern peninsular Italy, Sicily, and the Maltese Islands (hereafter referred to as “ISM”) in order to detect population differentiation through the analysis of their molecular variance (AMOVA; Excoffier et al. Citation1992). Tajima’s D test and Fu’s FS test (Tajima Citation1989; Fu Citation1997) were performed in order to evaluate the population demography of this species. A mismatch distribution analysis was performed to estimate the parameters of demographic expansion. AMOVA, neutrality tests and mismatch analysis were performed with the software Arlequin (v. 3.5.2.2, Excoffier & Lischer Citation2010). In addition, haplotype diversity and nucleotide diversity were computed with the software DnaSP v. 5 (Librado & Rozas Citation2009). In order to estimate the significance of the pair-wise differences in haplotype diversity between the three geographical groups, we used a series of functions, developed by Thomas et al. (Citation2002) and available at http://www.ucl.ac.uk/mace-lab/resources/software (“TEST_h_DIFF”), through the statistical software “R” (v. 3.2.1, https://www.R-project.org ).
Past population demography was inferred using a Bayesian skyline plot model (Drummond et al. Citation2005), as implemented in BEAST v. 1.8.0 (Drummond et al. Citation2012) and visualised in TRACER v. 1.6 (Rambaut et al. Citation2014). The coalescent approach uses standard Monte Carlo Markov chain (MCMC) sampling procedures to estimate a posterior distribution of gene genealogies and population parameters under a TN93 + I substitution model (Tamura & Nei Citation1993). The choice of the correct evolutionary model was conducted using jModelTest (v. 2.0, Guindon & Gascuel Citation2003; Darriba et al. Citation2012) according to the Akaike information criterion (AIC; Akaike Citation1974). A log-normal model that relaxes the molecular clock hypothesis was used (Drummond et al. Citation2006). The hyper-parameter “m” (number of grouped intervals) was set to 10. Three independent MCMC analyses of 107 steps were performed, sampling every 10,000th generation with the burn-in set at 106 generations. To calibrate the molecular clock, we used the same split as described by Jesse et al. (Citation2009), i.e. the one between Potamon fluviatile and P. algeriense, which most likely took place at the end of the Messinian salinity crisis (Brandis et al. Citation2000).
Results
The obtained sequences aligned unambiguously and translated without stop codons, thus suggesting that no pseudogenes were erroneously amplified. A total of 11 haplotypes, including both the haplotypes observed by Jesse et al. (Citation2009) and two new haplotypes recorded within the framework of the current study (“NH11” and “NH10”; GenBank accession numbers KY449461 and KY449462; ), were obtained. The three geographical groups showed the following haplotype diversity values (Hd): 0.681 (Balkans), 0.040 (Central Italy) and 0.527 (ISM). The following nucleotide diversity (Pi) values were obtained: 0.0042 for the group “Balkans”, 0.0008 for the group “Central Italy”, and 0.0298 for “ISM”. In terms of pair-wise haplotype diversity differences between the three geographical groups, we obtained significant p values (< 0.0001), whereas the comparison between the Balkan and the ISM cohorts was insignificant (p value = 0.057). The haplotype network obtained shows that there are two common and widespread haplotypes shared by all three groups, plus some rarer haplotypes shared by a single or by a few individuals which are restricted to single areas (). The AMOVA test () shows that the percentage of variation between the three geographical groups is 41.90% while the variation within the groups is 58.10%, with an Fixation index (FST) value of 0.419 and a p value < 0.0001.
Table II. AMOVA and FST results based on 502-bp-long fragments of the mtDNA COI gene.
The neutrality tests are not significant for the “Balkans” and “ISM” groups, whilst the Fu’s FS statistics p value is only significant for the group “Central Italy”. Specifically, Tajima’s D has a value of −1.185 for “Balkans”, −1.100 for “Central Italy” and 0.367 for “ISM”. Conversely, Fu’s FS values of −2.736 for the “Balkans” and of −1.652 for the “Central Italy” group were recorded, with a p value and a corresponding Fu’s FS value of 0.037 and 1.246, respectively, for the “ISM” group ().
Table III. Neutrality test results based on 502-bp-long fragments of the mtDNA COI gene.
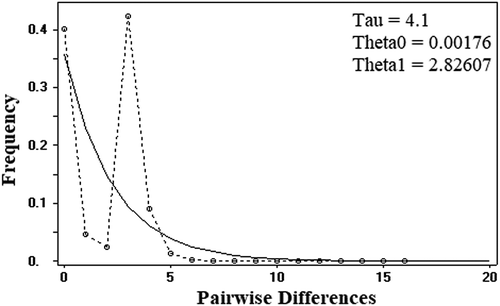
The mismatch analysis carried out on the mtDNA COI sequences showed a skewed unimodal curve with the observed number of pair-wise differences, which differs non-significantly from the expected values (Sum of square deviations (SSD) p value = 0.020; raggedness index (RI) = 0.317; raggedness p value = 0.010; ).
Figure 3. Result of the mismatch distribution analysis based on a 502-bp-long fragment of the mitochondrial DNA gene Cytochrome oxidase subunit 1 (COI). The dotted line represents the observed frequency distribution of pairwise differences; the curve represents the expected one.
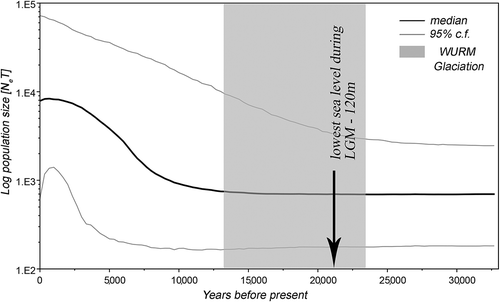
The estimated change in effective population size over time with 95% confidence intervals as computed with the Bayesian skyline plot analysis (BSP) is shown in . As indicated by the BSP analysis, the demographic expansion began approximately 12,000 years before present (BP), with a 95% confidence interval ranging from 6000 to 24,000 years BP.
Discussion
No apparent associations between observed haplotypes and their geographic locations were found, i.e. individual haplotypes could not be ascribed to exclusive geographical locations. However, the haplotype diversity values observed for the three geographical groups indicate the occurrence of a highly significant differentiation for the pairs “Balkans vs. Central Italy” and “ISM vs. Central Italy”; conversely, no significant difference was scored for the pair “Balkans vs. ISM”.
The performed analyses show that the studied Potamon fluviatile populations are not geographically structured, and that the highest haplotype diversity is present in the Balkan area, despite the relatively low number of samples studied from this region. Our results are thus in agreement with the hypothesis formulated by Jesse et al. (Citation2009) that, during the Pleistocene, southern Italy and the Maltese Islands did not act as refugia where early Pleistocene or pre-Quaternary P. fluviatile local populations survived the glaciation periods, but rather they proved to be a “land of conquest” for the Balkan P. fluviatile populations, which could colonise these new lands made accessible by virtue of the marine regressions linked with Pleistocene glaciations. Such a hypothesised dispersal pattern finds a counterpart in other freshwater crustaceans, such as in the anostracan Chirocephalus kerkyrensis Pesta, 1921, whose demographic expansions consistently coincided with the interglacial warm phases of the Pleistocene, while its spatial expansion (and colonisation of Italy from a Balkan origin) was restricted to the Pleistocene cold periods (Ketmaier et al. Citation2012). Later on, at the end of the Pleistocene, when temperatures rose gradually, Potamon fluviatile might have actively colonised more central and northern regions of Italy from its southern enclaves situated along the same peninsula. This secondary northward migration of the species was denoted by relatively low haplotype diversity values in analysed central Italy populations, despite the relatively high number of sampled individuals from this region, which in turn might be symptomatic of a founder effect.
The existence of a recent genetic exchange between the different populations within the studied areas (or of recent colonisation events via extensive dispersal with no founder effect) is suggested by the AMOVA test results. In the light of the FST values obtained and the results from the mismatch analysis, the species is inferred to be in a process of ongoing demographic expansion.
In the Bayesian skyline plot analysis, the upper 95% confidence interval suggests that this inferred demographic expansion could have started even before the last glacial maximum (LGM), when the freshwater crab is assumed to have reached southern peninsular Italy, and subsequently Sicily and the Maltese archipelago, through a trans-Adriatic route (Jesse et al. Citation2009). Moreover, considering that there is evidence of maritime activities in the Western Mediterranean dating back to 6000 years BP (Broodbank Citation2006 in Jesse et al. Citation2009), from the lower 95% confidence interval we cannot exclude the possibility of an anthropogenic introduction of Potamon fluviatile in peninsular Italy, which would thus be an example of “cryptic invasion” (e.g. Marrone et al. Citation2011). However, areas recently colonised through a human-mediated introduction are usually characterised by substantially lower haplotype richness values when compared, for example, to the haplotype richness values observed in the current study for the group “ISM”. Consequently, and in accordance with Jesse et al. (Citation2009), we consider the anthropogenic introduction of the species in Italy and Malta as rather unlikely.
In the light of the likely autochthonous status of the species throughout its current distribution range, a renewed conservation effort aimed at maintaining the populations of this freshwater decapod should be made, especially since they are currently facing multiple pressures (Gherardi & Holdich Citation1999; Füreder et al. Citation2002; Svoboda et al. Citation2014).
Table S1
Download MS Word (21.5 KB)Acknowledgements
This study was funded by the Research Committee of the University of Malta, to which the authors are greatly indebted. The Environment and Resources Authority (ERA) of Malta is acknowledged for having provided the sampling permit to AD (File ref.: NP 27/15) authorising the sampling of Potamon fluviatile individuals within four Maltese sites. Francesco Sacco (Palermo, Italy) is acknowledged for the support he provided.
Supplemental data
Supplemental data for this article can be accessed here
Additional information
Funding
References
- Akaike H. 1974. A new look at the statistical model identification. IEEE Transactions on Automatic Control 19:716–723. DOI: 10.1109/TAC.1974.1100705.
- Bandelt H , Forster P , Röhl A. 1999. Median-joining networks for inferring intraspecific phylogenies. Molecular Biology and Evolution 16:37–48. DOI: 10.1093/oxfordjournals.molbev.a026036.
- Barbaresi S , Cannicci S , Vannini M , Fratini S . 2007. Environmental correlates of two macro-decapods distribution in Central Italy: Multi-dimensional ecological knowledge as a tool for conservation of endangered species. Biological Conservation 136:431–441. DOI: 10.1016/j.biocon.2006.12.013.
- Bezerra AMR , Annesi F , Aloise G , Amori G , Giustini L , Castiglia R . 2015. Integrative taxonomy of the Italian pine voles, Microtus savii group (Cricetidae, Arvicolinae). Zoologica Scripta 45:1463–6409. DOI: 10.1111/zsc.12155.
- Bott R . 1967. Potamidae (Crustacea, Decapoda) aus Afghanistan, Westasien und dem Mittelmeerraum (Eine Revision der Untergattung Potamon s str.). Videnskabelige Meddelelser Dansk Naturhistorisk Forening 130:7–43.
- Brandis D , Storch V , Turkay M . 2000. Taxonomy and zoo-geography of the freshwater crabs of Europe, North Africa, and the Middle East. Senckenbergiana Biologica 80:5–56.
- Broodbank C . 2006. The origins and early development of Mediterranean maritime activity. Journal of Mediterranean Archaeology 19:199–230. DOI: 10.1558//jmea.2006.v19i2.199.
- Canestrelli D , Cimmaruta R , Nascetti G . 2007. Phylogeography and historical demography of the Italian treefrog, Hyla intermedia, reveals multiple refugia, population expansions and secondary contacts within peninsular Italy. Molecular Ecology 16:4808–4821. DOI: 10.1111/j.1365-294X.2007.03534.x.
- Canestrelli D , Nascetti G . 2008. Phylogeography of the pool frog Rana (Pelophylax) lessonae in the Italian peninsula and Sicily: Multiple refugia, glacial expansions and nuclear-mitochondrial discordance. Journal of Biogeography 35:1923–1936. DOI: 10.1111/j.1365-2699.2008.01946.x.
- Cumberlidge N 2008. Potamon fluviatile. The IUCN Red List of Threatened Species 2008:E.T134293A3933275. Available: http://dx.doi.org/10.2305/IUCN.UK.2008.RLTS.T134293A3933275.en . Accessed May 2015 20.
- Darriba D , Taboada GL , Doallo R , Posada D . 2012. jModelTest 2: More models, new heuristics and parallel computing. Nature Methods 9:772. DOI: 10.1038/nmeth.2109.
- Das S . 2015. Morphological, molecular, and hormonal basis of limb regeneration across Pancrustacea. Integrative and Comparative Biology 55:869–877. DOI: 10.1093/icb/icv101.
- Drummond AJ , Ho SYW , Phillips MJ , Rambaut A . 2006. Relaxed phylogenetics and dating with confidence. PLoS Biology 4:699–710. DOI: 10.1371/journal.pbio.0040088.
- Drummond AJ , Rambaut A , Shapiro B , Pybus OG . 2005. Bayesian coalescent inference of past population dynamics from molecular sequences. Molecular Biology and Evolution 22:1185–1192. DOI: 10.1093/molbev/msi103.
- Drummond AJ , Suchard MA , Xie D , Rambaut A . 2012. Bayesian phylogenetics with BEAUti and the BEAST v. 1.7. Molecular Biology and Evolution 29:1969–1973. DOI: 10.1093/molbev/mss075.
- Excoffier L , Lischer HEL . 2010. Arlequin suite v. 3.5: A new series of programs to perform population genetics analyses under Linux and Windows. Molecular Ecology Resources 10:564–567. DOI: 10.1111/j.1755-0998.2010.02847.x.
- Excoffier L , Smouse PE , Quattro JM . 1992. Analysis of molecular variance inferred from metric distances among DNA haplotypes: Application to human mitochondrial DNA restriction data. Genetics 131:479–491.
- Fu YX . 1997. Statistical tests of neutrality of mutations against population growth, hitchhiking, and background selection. Genetics 147:915–925.
- Füreder L , Oberkofler B , Hanel R , Machino Y . 2002. Freshwater crayfish in South Tyrol (Italy): Distribution and protection measures of endangered Austropotamobius pallipes . Bulletin Français de la Pêche et de la Pisciculture 367:651–662. DOI: 10.1051/kmae:2002056.
- Gherardi F , Guidi S , Vannini M . 1987. Behavioural ecology of the freshwater crab, Potamon fluviatile: Preliminary observations. Investigacion Pesquera 51:389–402.
- Gherardi F , Holdich DM, editors . 1999. Crayfish in Europe as alien species. In: How to make the best of a bad situation? Rotterdam: A.A. Balkema.
- Guindon S , Gascuel O . 2003. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Systematic Biology 52:696–704. DOI: 10.1080/10635150390235520.
- Herbst JWF . 1785. Versuch einer naturgeschichte der krabben und krebse. Johann Caspar Fuessly, Zurich 1:183–206.
- Hewitt G . 2000. The genetic legacy of the Quaternary ice ages. Nature 405:907–913. DOI: 10.1038/35016000.
- Hewitt G . 2004. Genetic consequences of climatic oscillations in the Quaternary. Philosophical Transactions of the Royal Society of London Series B 359:183–195. DOI: 10.1098/rstb.2003.1388.
- Jesse R , Grundiski M , Klaus S , Streit B , Pfenninger M . 2011. Evolution of freshwater crab diversity in the Aegean region (Crustacea: Brachyura: Potamidae). Molecular Phylogenetics and Evolution 59:23–33. DOI: 10.1016/j.ympev.2010.12.011.
- Jesse R , Pfenninger M , Fratini S , Scalici M , Streit B , Schubart CD . 2009. Disjunct distribution of the Mediterranean freshwater crab Potamon fluviatile – Natural expansion or human introduction? Biological Invasions 11:2209–2221. DOI: 10.1007/s10530-008-9377-0.
- Jesse R , Schubart CD , Klaus S . 2010 . Identification of a cryptic lineage within Potamon fluviatile (Herbst, 1785) (Crustacea: Brachyura: Potamidae). Invertebrate Systematics 24:348–356. DOI: 10.1071/IS10014.
- Ketmaier V , Marrone F , Alfonso G , Paulus K , Wiemann A , Tiedemann R , Mura G . 2012. Mitochondrial DNA regionalism and historical demography in the extant populations of Chirocephalus kerkyrensis (Branchiopoda: Anostraca). PLOS One 7:e30082. DOI: 10.1371/journal.pone.0030082.
- Kindler C , Bohme W , Corti C , Gvozdik V , Jablonski D , Jandzik D , Metallinou M , Siroky P , Fritz U . 2013. Mitochondrial phylogeography, contact zones and taxonomy of grass snakes (Natrix natrix, N. megalocephala). Zoologica Scripta 42:458–472. DOI: 10.1111/zsc.12018.
- Kumar S , Stecher G , Tamura K . 2016. MEGA7: Molecular Evolutionary Genetics Analysis version 7.0 for bigger datasets. Molecular Biology and Evolution 33:1870–1874. DOI: 10.1093/molbev/msw054.
- Larkin MA , Blackshields G , Brown NP , Chenna R , McGettigan PA , McWilliam H , Valentin F , Wallace IM , Wilm A , Lopez R , Thompson JD , Gibson TJ , Higgins DG . 2007. Clustal W and Clustal X v. 2.0. Bioinformatics 23:2947–2948. DOI: 10.1093/bioinformatics/btm404.
- Librado P , Rozas J . 2009. DnaSP v. 5: A software for comprehensive analysis of DNA polymorphism data. Bioinformatics 25:1451–1452. DOI: 10.1093/bioinformatics/btp187.
- Marrone F , Lo Brutto S , Arculeo M . 2011. Cryptic invasion in Southern Europe: The case of Ferrissia fragilis (Pulmonata: Ancylidae) Mediterranean populations. Biologia - Section Zoology 66:484–490. DOI: 10.2478/s11756-011-0044-z.
- Matthew MA , Reynolds JD . 1995. A population study of the white-clawed crayfish Austropotamobius pallipes (Lereboullet) in an Irish reservoir. Biology Environment: Proceedings of the Royal Irish Academy 95B:99–109.
- Mazza G , Tricarico E , Cianferoni F , Stasolla G , Inghilesi AF , Zoccola A , Innocenti G . 2017. Native crab and crayfish co-occurrence: First evidence in Europe. Biologia 72:790–795. DOI: 10.1515/biolog-2017-0086.
- Michaux JR , Magnanou E , Paradis E , Nieberding C , Libois R . 2003. Mitochondrial phylogeography of the Woodmouse (Apodemus sylvaticus) in the Western Palearctic region. Molecular Ecology 12:685–697. DOI: 10.1046/j.1365-294X.2003.01752.x.
- Micheli F , Gherardi F , Vannini M . 1990. Growth and reproduction in the freshwater crab, Potamon fluviatile (Decapoda, Brachyura). Freshwater Biology 23:491–503. DOI: 10.1111/j.1365-2427.1990.tb00290.
- Rambaut A , Suchard MA , Xie D , Drummond AJ 2014. Tracer v. 1.6. Available: http://beast.bio.ed.ac.uk/Tracer . Accessed May 2017 28.
- Richterich P . 1998. Estimation of errors in “raw” DNA sequences: A validation study. Genome Research 8:251–259. DOI: 10.1101/gr.8.3.251.
- Scalici M , Goretti E , Dörr AJM , D’Allestro V . 2013. Morphometric homogeneity and allometric growth of Potamon fluviatile in Italy. Crustaceana 86:1291–1296. DOI: 10.1163/15685403-00003234.
- Schubart CD , Huber MGJ . 2006. Genetic comparisons of German populations of the stone crayfish, Austropotamobius torrentium (Crustacea: Astacidae). Bulletin français de la pêche et de la pisciculture 380–381:1019–1028. DOI: 10.1051/kmae:2006008.
- Svoboda J , Strand DA , Vrålstad T , Grandjean F , Edsman L , Kozák P , Kouba A , Fristad RF , Bahadir Koca S , Petrusek A . 2014. The crayfish plague pathogen can infect freshwater-inhabiting crabs. Freshwater Biology 59:918–929. DOI: 10.1111/fwb.12315.
- Tajima F . 1989. Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics 123:585–595.
- Tamura K , Nei M . 1993. Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Molecular Biology and Evolution 10:512–526.
- Thomas MG , Weale ME , Jones AL , Richards M , Smith A , Redhead N , Torroni A , Scozzari R , Gratrix F , Tarekegn A , Wilson JF , Capelli C , Bradman N , Goldstein DB . 2002. Founding mothers of Jewish communities: Geographically separated Jewish groups were independently founded by very few female ancestors. American Journal of Human Genetics 70:1411–1420. DOI: 10.1086/340609.
- Vamberger M , Stuckas H , Sacco F , D’Angelo S , Arculeo M , Cheylan M , Corti C , Lo Valvo M , Marrone F , Wink M , Fritz U . 2015. Differences in gene flow in a twofold secondary contact zone of pond turtles in southern Italy (Testudines: Emydidae: Emys orbicularis galloitalica, E. o. hellenica, E. trinacris). Zoologica Scripta 44:233–249. DOI: 10.1111/zsc.12102.
- Yeo D , Ng P . 2003. Recognition of two subfamilies in the Potamidae Ortmann, 1896 (Brachyura, Potamidae) with a note on the genus Potamon Savigny, 1816. Crustaceana 76:1219–1235. DOI: 10.1163/156854003773123456.