Abstract
The Rosalia longicorn (Rosalia alpina) is a strictly protected saproxylic beetle, widely distributed in Central and Southern Europe and mainly associated with ancient beech forests. To improve knowledge about the conservation status of R. alpina in Italy, available molecular markers (microsatellites and mitochondrial cytochrome c oxidase I(COI)) were tested for the first time on Italian populations. The study was performed in four sampling sites distributed in two areas placed in Northern (“Foreste Casentinesi” National Park) and Central Apennines (“Abruzzo, Lazio and Molise” National Park) where populational data about Rosalia longicorn were collected in the framework of the European LIFE MIPP Project. The genetic relationship among Apennine and Central/South-eastern European populations was explored by a comparison with mitochondrial DNA (mtDNA) data from literature. Microsatellite markers were only partially informative when applied to R. alpina Italian individuals, although providing some preliminary indication on an extensive gene flow among populations from the Apennines and local ongoing processes of genetic erosion. Genetic data are consistent with previous ecological data suggesting that the maintenance of variability in this species could be related to both habitat continuity and preservation of large senescent or standing dead trees in forests. Finally, a peculiar origin of the Apennine populations of R. alpina from a putative “Glacial Refugium” in Italy was inferred through COI data. The high genetic distance scored among the analysed populations and those from Central and South-eastern Europe indicates that the R. alpina deme from Apennine Mountains might represent a relevant conservation unit in Europe. Further genetic analyses will allow assessing other possible conservation units of R. alpina and, thus, defining large-scale conservation strategies to protect this endangered longhorn beetle in Europe.
Introduction
The longhorn beetle Rosalia alpina (Linnaeus, 1758) (Coleoptera: Cerambycidae) is an obligate saproxylic species which depends on dead wood of dying and decaying trees, generally well spread in mature forests (Speight Citation1989; Alexander Citation2008; Stokland et al. Citation2012). The larvae of the Rosalia longicorn complete their development in 2–3 years, mainly feeding on deadwood of mature, dead (or moribund) and sun-exposed beech trees (Fagus sylvatica Linnaeus and F. orientalis Lipsky). Therefore, the habitat of the species consists in mountain forest clearings, wooded grasslands and forest patches, having a low percentage of canopy closure (Sama Citation1988, Citation2002; Duelli & Wermelinger Citation2005; Campanaro et al. Citation2017). It is worth noting that the larval stage can also develop in other deciduous trees (e.g. Ulmus, Acer, Carpinus, Tilia, Fraxinus, Castanea, Juglans, etc.) along a wide altitudinal range from the coastline to about 2000 m above sea level (asl; Švacha & Danilevsky Citation1988; Bense Citation1995; Ciach et al. Citation2007; Cizek et al. Citation2009; Di Santo & Biscaccianti Citation2014).
The Rosalia longicorn is widespread in Europe, mainly tracking the presence of the beech tree species, its main host plant, with a geographic range covering most of Central and Southern Europe from Asturias to the Ural Mountains (Müller Citation1950; von Demelt Citation1956; Sama Citation1988, Citation2002; Bense Citation1995; Bense et al. Citation2003; but see also the distributional map outlined in of Drag et al. Citation2015). The northern limit of its current distribution runs through North-western Spain, France (up to the Seine), Southern Germany (Baden-Württemberg and Bavaria), the Czech Republic, Poland (western and southern territories), Southern Ukraine and the Southern Urals (Bense Citation1995; Shapovalov Citation2012; Michalcewicz & Ciach Citation2015); old records, not yet confirmed, exist for Northern Germany, Denmark and Southern Sweden (Lindhe et al. Citation2011). In Southern Europe, the species occurs in most mountain areas of Italy and Corsica, where some relict populations also populate lowland forest fragments, but the core area of its distribution lies in South-eastern Europe and extends to the Caucasus through Northern Anatolia (Müller Citation1950; Von Demelt Citation1956; Sama Citation1988, Citation2002; Bense et al. Citation2003). The southern limit of the range runs through Northern Sicily (mainly Madonie and Nebrodi Mts.) and the Greek mainland, with a very isolated population in South-eastern Turkey (the western slope of the Nur Mts.), where an endemic subspecies was described, R. alpina syriaca Pic, 1895 (Sama Citation2002; Sama & Löbl Citation2010; Alì & Rapuzzi Citation2016).
Figure 1. Microsatellite locus RA_23 sequence alignment. Motif 1 and Motif 2 are highlighted in red and green, respectively. Reference sequence for RA_23 locus retrieved from GenBank, Acc. N° = KF114388.1 (Drag et al. Citation2015).

To better assess the conservation status of R. alpina and to plan conservation strategies at both fine and broad geographical scales, analysis of the genetic viability of this species is urgently required to supplement distributional and ecological data. To this aim, suitable molecular markers were developed (Drag et al. Citation2013) and have been used so far to investigate patterns of genetic structure among Central and South-eastern European populations (Drag et al. Citation2015). These studies highlighted that populations from Central Europe were genetically less variable than those of the Southern area, i.e. Northern-western Greece (Pindus range, Olympus Mts), where, likely, a glacial refugium occurred (Drag et al. Citation2015).
In Italy, R. alpina is mainly associated with ancient deciduous forests dominated by Fagus sylvatica (Lachat et al. Citation2013) in both the Alpine and Apennine ranges. The last assessment of the conservation status of this insect in the Italian peninsula (performed during the years 2006–2012) placed this species in the category “Inadequate (U1)” (Genovesi et al. Citation2014), and the recently published Italian Red List of Italian Saproxylic Beetles evaluated the species as “Nearly Threatened” (Audisio et al. Citation2014; Carpaneto et al. Citation2015). To perform a first genetic survey of Italian populations, we gathered data from R. alpina populations in two areas where monitoring protocols were recently standardized and performed in the framework of the EU-LIFE monitoring insects with public participation (MIPP) Project (Mason et al. Citation2015; Rossi De Gasperis Citation2016; Campanaro et al. Citation2017): the first area is placed in the Northern Apennines (“Foreste Casentinesi” National Park), and the second in the Central Apennines (“Abruzzo, Lazio and Molise” National Park).
With the present work we aimed at: (i) evaluating for the first time the performance of the available microsatellite markers (Drag et al. Citation2015) in Italian populations of R. alpina, and assessing their usefulness in detecting genetic structure at a fine geographical scale; (ii) estimating polymorphism levels in the two R. alpina Apennine populations and the gene flow between them, in order to provide hints about their current status of genetic conservation; and (iii) tracing the biogeographic origin of Italian populations by exploring the relationship among mitochondrial DNA lineages of the Apennines and those from Central and South-eastern Europe (Drag et al. Citation2015).
Material and methods
Study areas, sample collection and DNA extraction
Collection of individuals was performed during the monitoring of R. alpina carried out in the framework of the EU LIFE-MIPP Project (Campanaro et al. Citation2017; Carpaneto et al. Citation2017; Rossi De Gasperis et al. Citation2017) in two Italian National Parks (also “Sites of Community Importance” or SCI): “Foreste Casentinesi, Monte Faltenora e Campigna” National Park (hereafter abbreviated as “FC”; SCI: IT4080001), located in the Northern Apennines (Forlì-Cesena and Arezzo provinces), and “Abruzzo Lazio e Molise” National Park (hereafter abbreviated as “PA”; SCI: IT7110205), located in the Central Apennines (L’Aquila, Isernia and Frosinone provinces). The distance between the two areas is about 250 km. The beetles were captured and handled under a permit from the Italian Ministry of Environment (Prot. 0044591/PNM 16/09/2013).
The study area within the FC corresponded to the locality “Foresta della Lama” (700 m asl; 43.4312°N, 11.8381°E), characterized by closed forests dominated by Fagus sylvatica and Abies alba. In PA, three sub-areas were selected: “Difesa di Pescasseroli” (DP; 1300 m asl; 41.8461°N, 13.8600°E), and “Val Fondillo” (VF; 1200 m asl; 41.7841°N, 13.9563°E), characterized by pure beech forests with wide clearings and old growth trees (the distance between these two sub-areas is about 6.5 km); and “Zio Mas” (ZM; 1700 m asl; 42.0802°N, 14.0566°E), characterized by fragmented beech woodlands interspersed among open mountain grasslands (distant ~10 and 30 km from VF and DP, respectively; for more detail on the ecological features of the sampling areas see Carpaneto et al. Citation2017). The sample tissues for molecular analysis were obtained over a 2-year sampling period (2014–2015, during July and August), from specimens caught on trees along specific sampling transects (for details on positions of “wild trees” and the areas covered by the sampling transects, see figs 6 and 7 in Campanaro et al. Citation2017). Tarsomeres of one middle leg of the insect (as in Drag et al. Citation2015) were stored in vials containing 96% ethanol. DNA was extracted from a total of 89 specimens using the Genomic DNA Mini Kit Tissue (Geneaid) following the manufacturer’s instructions, but performing a longer (12 h) cell-lysis step.
Microsatellites: amplification, fragment size detection and data analysis
Eight of the nine microsatellite loci available for R. alpina (Drag et al. Citation2013) were amplified: RA_08, RA_11, RA_13, RA_15, RA_23, RA_28, RA_37 and RA_40 (loci alternatively labelled with 6-FAM or HEX). RA_29 was excluded a priori, since failure in DNA amplification at this locus was already reported (Drag et al. Citation2015). Microsatellite amplification protocols were optimized by a “touchdown” annealing procedure (Don et al. Citation1991) and extension of the final elongation step to 15 min to ensure complete polyadenylation of DNA strands. Polymerase chain reaction (PCR) thermal conditions were as follows: 4 min of initial denaturation at 94°C, followed by 15 cycles of 94°C (30 sec), 58 to 54°C (60 sec) by decreasing 0.2°C/cycle, 72°C (60 sec), followed by 25 cycles of 94°C (30 sec), 54°C (60 sec) and 72°C (60 sec), with final elongation at 72°C for 15 min. PCR fragment lengths were analysed with an ABI 3730XL automated sequencer (Applied Biosystems) by Macrogen Europe (The Netherlands). GeneMarker 2.6.3 (SoftGenetics LLC®) was used to evaluate microsatellite peak quality. Semi-automated selection of fragment-length polymorphisms at each locus was performed with STRand Analysis Software 2.04.0059 (Toonen & Hughes Citation2001). Frequencies of null alleles were estimated with Micro-Checker 2.2.3 (Van Oosterhout et al. Citation2004). GenAlEx 6.502 (Peakall & Smouse Citation2012) was used to calculate the observed (HO) and expected (HE) heterozygosity, the number of alleles per locus (A), the number of private alleles (AP), the allele frequencies (AF), the inbreeding coefficient (FIS), the fixation index (FST) and the multi-locus matches (GM) (i.e. to detect unique or shared multi-locus genotypes in populations). Computation of allelic richness (AR) was performed with Fstat 2.9.3.2 (Goudet Citation2002). Hardy–Weinberg equilibrium (HWE) at each locus/population was tested both for “heterozygote deficiency” and for “heterozygote excess” through the Markov chain algorithms (MC) using the default parameters set on Genepop 4.2 (Rousset Citation2008). Significant genotypic differentiation (GD) among populations was detected with Genepop 4.2.
COI: amplification, sequencing and data analysis
A partial fragment of the mitochondrial cytochrome c oxidase subunit I (COI, ~760 bp) was amplified on a sub-sample of 36 individuals (N = 13 in FC; N = 23 in PA). Amplifications were carried out using universal primers (F: C1-J-2183; R: TL2-N-3014; Simon et al. Citation1994) and thermal conditions reported in Drag et al. (Citation2015). Sequencing of PCR products was performed by Macrogen Europe (The Netherlands). Quality of COI chromatograms was assessed by PREGAP4 and GAP4 software implemented in the Staden Package 2.0.0 (Staden et al. Citation1998). COI sequences (deposited in GenBank under accession numbers MG930944-MG930979) were aligned with those already available in GenBank from other European R. alpina populations (Drag et al. Citation2015). Genetic polymorphism parameters, such as number of haplotypes, haplotype diversity (h), nucleotide diversity (π) and Tajima’s D (Tajima Citation1989) were computed using DnaSP v5.10 (Librado & Rozas Citation2009). Relationship among haplotypes were built in a phylogenetic network produced using the Statistical Parsimony method (TCS) (Clement et al. Citation2000) and depicted with the help of POPArt 1.7 (http://popart.otago.ac.nz).
Results
Microsatellites
Despite optimization, loci RA_11 and RA_15 failed to amplify in almost all samples, and, thus, these loci were excluded from subsequent analyses. Polymorphism levels for each amplified locus are reported in . Among loci, RA_23 appeared to be the most variable (14 alleles). However, after sequencing five homozygote individuals showing different fragment lengths (132–164 bp), RA_23 proved to be a “complex microsatellite” composed by at least two highly variable and different repeated motifs (“TG” and “GTCRTTGTC”; . Due to its complex nature, RA_23 was not further considered for polymorphism analysis. The mean number of alleles ranged from a monomorphic condition in RA_37 to a maximum of six alleles in RA_13 (). Thirteen percent null alleles was estimated in RA_40, in which one of the two alleles was rare (2.8%) and exclusive of DP. The FC population was in HWE over all loci ( and ), while a significant deviation from HWE was detected in the PA population (), with high (and significant) FIS, at both RA_13 and RA_40 (). Indeed, data indicated HWE for the three populations from PA sub-areas (i.e. DP, VF and ZM; ). Mean HE and AR values were comparable between Apennine populations from the two study sites, although both were slightly higher in PA (HE = 0.310 ± 0.119; AR = 2.49) than in FC (HE = 0.286 ± 0.119; AR = 2.20). In Abruzzo, the highest HE and AR were recorded in DP (HE = 0.323 ± 0.116; AR = 2.35), whereas the lowest values were observed in ZM (HE = 0.230 ± 0.117; AR = 2.00).
Table I. Microsatellite polymorphism at each locus. A = number of alleles; HO = observed heterozygosity; HE = expected heterozygosity.
Table II. Microsatellite polymorphism in Apennine Rosalia alpina populations. N = number of individuals; HO = mean observed heterozygosity; HE = mean expected heterozygosity; FIS = inbreeding coefficient (** p-value < 0.01; *** p-value < 0.001). FC = “Foreste Casentinesi” National Park; PA = “Abruzzo Lazio e Molise” National Park; ZM = “Zio Mas” sub-area; DP = “Difesa di Pescasseroli” sub-area; VF = “Val Fondillo” sub-area.
Table III. Microsatellite polymorphism at each locus in Apennine Rosalia alpina populations. HO = observed heterozygosity; HE = expected heterozygosity; FIS inbreeding coefficient (* p-value < 0.05; ** p-value < 0.01; *** p-value < 0.001). FC = “Foreste Casentinesi” National Park; PA = “Abruzzo Lazio e Molise” National Park; ZM = “Zio Mas” sub-area; DP = “Difesa di Pescasseroli” sub-area; VF = “Val Fondillo” sub-area.
The allele frequencies for polymorphic loci are reported in Figure 2. Four private alleles (one for each locus) were found in PA (frequencies = 2–8%). Among PA sub-areas, private alleles were observed in VF (RA_08, freq. = 0.063; coloured green in and in DP (RA_40, freq. = 0.074; coloured red in , but not in ZM. Among all loci, RA_13 (the most polymorphic) showed highly variable allele frequencies among populations (): a common allele for FC and PA was found (freq. > 0.6), while the other four alleles were equally frequent (~0.08) in FC, but not in PA, where an additional (and private) fifth allele was detected.
Figure 2. Pie chart of allele frequencies for the four polymorphic microsatellite loci in each Apennine Rosalia alpina population. Graphic representation of the mean allele number (A), mean allelic richness (AR) and mean number of private alleles (AP). Asterisks (*) indicate private alleles. Above the dashed line: comparison between “Foreste Casentinesi” and “Abruzzo Lazio e Molise” National Park populations; below the dashed line: comparison among populations from the three sub-areas of the “Abruzzo Lazio e Molise” National Park. FC = “Foreste Casentinesi”; PA = “Abruzzo Lazio e Molise” National Park; ZM = “Zio Mas” sub-area; DP = “Difesa di Pescasseroli” sub-area; VF = “Val Fondillo” sub-area.
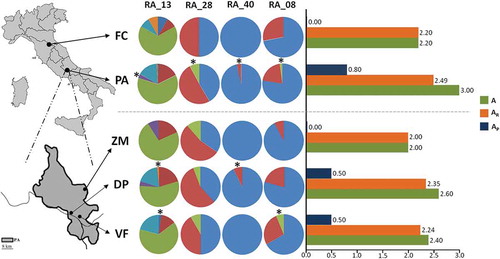
Overall, FC and PA populations were not significantly differentiated (FST = 0.009, p > 0.05). Comparisons among the three PA sub-areas highlighted a significant (but low) allelic (FST = 0.037, p < 0.05) and genotypic differentiation (GD; p = 0.023) between VF and ZM.
The multi-locus match analysis (also including genotype data from the “complex microsatellite” RA_23) showed that 88.8% of the analysed Apennine individuals had exclusive multi-locus genotypes, while 11.2% of them shared a multi-locus combination with only one other individual. When partitioning the data set into the two main Apennine populations, exclusive multi-locus genotypes were found in 88.7% and 100% of PA and FC individuals, respectively.
COI
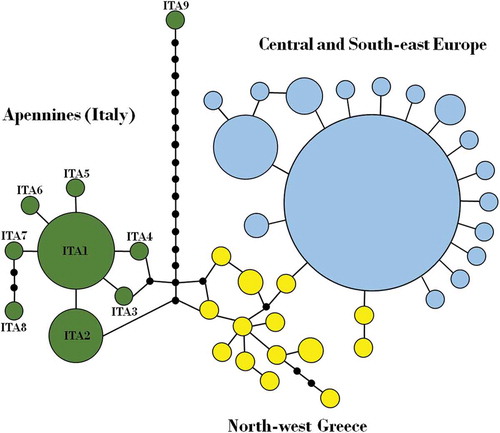
Nine COI haplotypes (h = 0.59; π = 0.23%) were detected for Apennine populations (Figure 3). No genetic structure was observed, but a higher nucleotide diversity was found in PA (π = 0.31%) than in FC (π = 0.10%). Two haplotypes were shared and more common in our sample (ITA1, freq. = 0.61; ITA2, freq. = 0.19), whereas the remaining seven haplotypes were unique for single FC (n = 3) and PA (n = 4) individuals. Haplotype ITA1 was only found in ZM. Haplotypes ITA8 (exclusive of VF) and ITA9 (exclusive of DP), showed four and 18 mutational steps from the most common haplotype (ITA1), respectively. Tajima’s D was negative (PA = −2.39; FC = −1.44) in both Apennine populations and significant only in PA (p < 0.01), indicating an excess of rare variants in this study area. All scored haplotypes proved to be peculiar to the Italian peninsula when compared with those observed in Central and South-eastern European populations (; Drag et al. Citation2015). Furthermore, Apennine haplotypes were more closely related to those from Northern Greece (e.g. two mutational steps from ITA2) than to other European haplotypes ().
Figure 3. TCS network showing relationship among cytochrome c oxidase I (COI) haplotypes from European Rosalia alpina populations. On the left, the nine Apennine COI haplotypes (in green); on the right, haplotypes from Central/South-eastern Europe (in blue) and North-west Greece (in yellow), modified from Drag et al. (Citation2015). Sizes of circles are proportional to haplotype frequencies; black dots indicate evolutionary mutational steps.
Discussion
Performance of microsatellite markers in Apennine populations of Rosalia alpina
Microsatellite markers available from the literature (Drag et al. Citation2013) proved to be only partially informative for estimating the genetic variability of Italian populations of R. alpina. In fact, two out of the eight analysed microsatellites (RA_11 and RA_15) failed to amplify in almost all Italian samples, probably because of point mutations in primer annealing sites. Microsatellite RA_23 seemingly showed a very high polymorphism (14 alleles), but sequencing revealed a “complex” mutation pattern (i.e. the presence of more than one repeat motif at this single locus), that strongly hinders the application of this specific marker in evolutionary analyses (). Finally, RA_37 was not informative at all, as it was found to be monomorphic in all analysed individuals. The remaining four microsatellites (RA_08; RA_13; RA_28; RA_40) showed a moderate to high polymorphism and, thus, provided some preliminary information on R. alpina genetic variability in Apennine populations.
Genetic structure of Rosalia alpina populations in the study areas
Microsatellite analysis did not indicate any substantial shared genetic structure between the two investigated Apennine populations of R. alpina (PA and FC). This could be explained assuming an historical gene flow among R. alpina populations from these two Apennine sectors, probably favoured by a rapid expansion of beech forests in the post-glacial period of the Middle Pleistocene (Magri et al. Citation2006) which ensured a continuity of suitable habitat for the expansion of this species. The absence of a substantial genetic differentiation between the two Apennine populations is also consistent with previous studies suggesting a high vagility of this beetle species (Russo et al. Citation2010; Bosso et al. Citation2013; Drag et al. Citation2011, Citation2015; Campanaro et al. Citation2017; Rossi De Gasperis et al. Citation2017). Indeed, R. alpina individuals can fly for long distances (i.e. more than 1.5 km; Drag et al. Citation2011; Rossi De Gasperis et al. Citation2017) although they usually move within a habitat patch in the range of dozens or hundreds of metres (Drag et al. Citation2011). However, the current habitat of this species is highly fragmented in Italy, with an average gap-distance of about 15 km that could be sufficient to represent an obstacle to the movement of individuals (Bosso et al. Citation2013). Hence, Italian populations of R. alpina might be currently particularly prone to a geographic isolation, leading to a reduction of gene flow and loss of diversity by genetic drift. The lower genetic polymorphism (in terms of both allele number and heterozygosity; and ) found in the population from FC in comparison to those from PA might reflect an ongoing reduction in the size of the effective population inhabiting the former area. Indeed, suitable habitats constituted by large decaying trees in opened areas (forest clearings to wooded grasslands) are less abundant in FC (Rossi De Gasperis Citation2016), and this might have represented a limiting factor for the population size of R. alpina in this study site. However, because of the scarce number of captured individuals in FC (Campanaro et al. Citation2017), a more extensive genetic survey in this area should be performed to confirm the observed pattern. In PA, the lowest genetic diversity (in both allelic richness and heterozygosity) was observed in the R. alpina population from the locality ZM, which also appeared genetically isolated from that of VF (but not significantly isolated from DP). This population is in fact more distant from and less contiguous with the other two sub-areas (), and inhabits a locally “patchy” habitat characterized by fragmented beech woodlands interspersed with open mountain grasslands (Rossi De Gasperis Citation2016; Campanaro et al. Citation2017). Furthermore, in ZM there are less suitable trees for R. alpina colonization (i.e. decaying trees in an advanced state of decomposition with trunk diameters larger than 100 cm) than in VF and DP (Rossi De Gasperis Citation2016). Geographic isolation and loss of suitable habitats might have contributed to trigger a process of genetic erosion in ZM sub-population, but the analysis of a larger number of individuals would allow greater statistical power to exclude possible sampling issues.
Comparison of mitochondrial patterns among Italian and Central/South-eastern European populations of Rosalia alpina
The presence of exclusive mitochondrial (mtDNA) haplotypes in the Italian samples suggests that the Apennine populations of R. alpina are highly differentiated from those of Central and South-eastern Europe. This genetic pattern recalls the isolation of North-western Greece populations (Pindus range, Olympus Mts) of R. alpina from those of Central Europe (Drag et al. Citation2015) and is a likely consequence of palaeoecological events that shaped the genetic variability of this species. The biogeographical history of R. alpina could overlap, at least in part, with that of its main host plant F. sylvatica, which survived in Europe during the Last Glacial period in multiple refugial areas (Magri et al. Citation2006; Magri Citation2008). Northern Greece and South-central Italian refugia for beech forests were likely isolated from those of Central Europe, and gave origin to three main genetic lineages that colonized the European continent (Magri et al. Citation2006; Magri Citation2008). The presence of the three main mitochondrial lineages of R. alpina in Europe, in line with those detected in F. sylvatica by chloroplast markers (Magri et al. Citation2006), might reflect the expansion of this beetle in Europe from different glacial refugia, following the beech forest during re-colonization. The highest mtDNA affinity observed among Apennine and North-west Greek R. alpina populations reflects the peculiar genetic similarity observed between Apennine and Balkan populations of F. sylvatica (as suggested by nuclear markers; Magri et al. Citation2006; Magri Citation2008) and could be related to the biogeographic history of the beech. The partial regression of the Adriatic Sea during the Last Glacial period would have caused a connection between the Northern and Central Italy and Balkan Peninsula (Pilaar Birch & Vander Linden Citation2017), which promoted the exchange of biota between the two peninsulas. Although this scenario should be investigated with further analysis, the same palaeogeographic scenario has been proposed to explain the high genetic affinity between Greece and Southern Italian populations of Morimus asper Sulzer, 1776 (Coleoptera, Cerambycidae) (Solano et al. Citation2013), a flightless saproxylic beetle inhabiting deciduous forests.
Finally, the higher mtDNA haplotype diversity (as well as the higher polymorphism found in microsatellites) found in PA than in FC, could be consistent with the typical pattern of “southern richness vs. northern purity” (Hewitt Citation2000). This greater genetic richness is usually attributed to the prolonged population stability of temperate species in southern refugia coupled with the loss of variation during post-glacial northward re-colonization. Hence, Apennine populations of R. alpina could have originated through northward dispersal and expansion from ancestral populations confined to glacial refugia of Southern and Central Italy (as also suggested by the negative Tajima’s D). The lower genetic variability observed in FC could be related to the longer distance from this site in the Northern Apennines, with respect to the PA site, from a putative glacial refugium in Central or Southern Italy.
Remarks on conservation of Rosalia alpina in Italy
The investigated Apennine R. alpina populations appeared genetically distinct from all other European populations and, therefore, these might represent important conservation units for Europe. Our data suggest that the effects of inbreeding can be negligible in the analysed populations (). The high percentage of exclusive multi-locus genotypes in Apennine populations also points to the occurrence of outcrossing (and, thus, recombination) redistributing the available genetic diversity into novel genotypic combinations. The high nucleotide diversity and presence of peculiar mtDNA haplotypes in both VF and DP suggest that the effective population sizes of these two populations may be sufficient to maintain an adequate level of genetic variability in these two sub-areas of the PA site. However, the lower heterozygosity and allelic richness found in ZM and in FC may represent the first hints of an ongoing process of loss of genetic variability in these two populations. Although the low genetic variability in FC could also be explained with the long distance of this site from a putative “glacial refugium” in Central or Southern Italy (Hewitt Citation2000), our data do not allow us to definitively rule out the alternative hypothesis. However, the maintenance of genetic variability in R. alpina seems to be strongly related to both habitat continuity and preservation. Finally, it is worth remarking that R. alpina has a long larval developing time (2–3 years) that contributes to lowering the intrinsic genetic polymorphism of the species, especially at the nuclear genome, as in other protected saproxylic beetles, i.e. M. asper (Solano et al. Citation2013) and the Osmoderma eremita (Scopoli, 1763) (Coleoptera, Scarabeidae) complex (Oleksa et al. Citation2013; Zauli et al. Citation2016).
Rosalia alpina might be more prone to rapid genetic erosion in the future if special protection measures are not assured. The most important management strategy to protect R. alpina and promote gene flow among populations is the preservation of “key trees” (large senescent or standing dead trees), on which mating and oviposition occur more frequently (Campanaro et al. Citation2017). Other effective actions would be to pollard the tree branches to reduce the risk of collapse of trees and expose their trunks to sunlight (Castro & Fernández Citation2016), and to avoid the stacking of timber in woodpiles that could act as attractive traps during oviposition, as for other saproxylic beetles (Ilić & Ćurčić Citation2013; Lachat et al. Citation2013). In addition, monitoring (for instance, through a non-invasive photographic identification method: Rossi De Gasperis et al. Citation2017) of populations will be necessary to assess the effectiveness of the implemented conservation plans in natural areas.
Novel and additional molecular markers, as well as further genetic analyses, will be necessary in the future to investigate gene flow among R. alpina populations at fine and large geographic scales and to assess their conservation status. Gathering additional genetic data from other Italian and European populations will also allow a better understanding of their phylogeographic relationship and possibly highlight other conservation units of R. alpina over its distributional range.
Acknowledgements
The present work was developed during the MS thesis of the first author (Marco Molfini), within the EU project LIFE11 NAT/IT/000252 (MIPP), with the contribution of the LIFE financial instrument of the European Union. The authors thank all the MIPP staff and the institutions involved: Comando Unità Tutela Forestale Ambientale ed Agroalimentare Carabinieri; Consiglio per la Ricerca in Agricoltura e l’Analisi dell’Economia Agraria – Difesa e Certificazione; Sapienza – Università degli Studi di Roma, Dipartimento di Biologia e Biotecnologie; Università degli Studi Roma Tre, Dipartimento di Scienze; and Ministero dell’Ambiente e della Tutela del Territorio e del Mare; Regione Lombardia. Moreover, the authors are grateful to the local offices of the Comando Unità Tutela Forestale Ambientale ed Agroalimentare Carabinieri that administrates the study sites: the UTCB Castel di Sangro (Tiziana Altea, Federica Desprini, Lucia Eusepi, Mario Romano) and the UTCB Pratovecchio (Silvia Bertinelli, Ester Giovannini, Angelo Lamberti, Sandro Aurelio Marsella, Valerio Mazzoli, Marcello Padula, Matteo Padula, Barbara Rossi, Giovanni Quilghini, Antonio Zoccola). We thank the staff of the Abruzzo, Lazio and Molise National Park (Carmelo Gentile, Cinzia Sulli, Paola Tollis) and the staff of the Foreste Casentinesi, Monte Falterona e Campigna National Park, and local operators (Cooperativa “In Quiete”, Cooperativa “Oros” and Guide ambientali escursionistiche “Quota 900”). We are grateful to all the field assistants who voluntarily helped us during the MIPP surveys. Thanks also to Daniela Lorenti and Pietro Mancini for assistance. We are especially grateful to Dr Verena Pichler for help in setting lab procedures and microsatellite data analysis, and to Dr Sönke Hardersen for comments on the manuscript and linguistic revision.
Additional information
Funding
References
- Alì K, Rapuzzi P. 2016. Second contribution to the knowledge of Longhorn Beetles of the Syrian Coastal Region (Coleoptera Cerambycidae). Biodiversity Journal 7:261–272.
- Alexander KNA. 2008. Tree biology and saproxylic Coleoptera: Issues of definitions and conservation language. Revue d’ Écologie 63:9–13.
- Audisio P, Baviera C, Carpaneto GM, Biscaccianti AB, Battistoni A, Teofili C, Rondinini C 2014. Lista Rossa IUCN dei Coleotteri saproxilici italiani. Comitato Italiano IUCN e Ministero dell‘Ambiente e della Tutela del Territorio e del Mare, Rome, Italy. [In Italian]
- Bense U. 1995. Longhorn beetles: Illustrated key to the cerambycidae and vesperidae of Europe. Weikersheim, Germany: Margraf.
- Bense U, Klausnitzer B, Bussler H, Schmidl J. 2003. Rosalia alpina (Linnaeus, 1758) Alpenbock. In: Petersen B, Ellwanger G, Biewald G, Hauke U, Ludwig G, Pretscher P, Schröder E, Ssymank A, editors. Das europäische Schutzgebietssystem Natura 2000. Ökologie und Verbreitung von Arten der FFH-Richtlinie in Deutschland, Schriftenreihe für Landschaftspflege und Naturschutz. Bonn: Bundesamt für Naturschutz. pp. 426−432.
- Bosso L, Rebelo H, Garonna AP, Russo D. 2013. Modelling geographic distribution and detecting conservation gaps in Italy for the threatened beetle Rosalia alpina. Journal for Nature Conservation 21:72–80.
- Campanaro A, Redolfi De Zan L, Antonini G, Hardersen S, Chiari S, Cini A, Mancini E, Mosconi F, Rossi de Gasperis S, Solano E, Bologna M, Sabbatini Peverieri G. 2017. Guidelines for the monitoring of Rosalia alpina. Nature Conservation 20:165–203.
- Carpaneto GM, Baviera C, Biscaccianti AB, Brandmayr P, Mazzei A, Mason F, Battistoni A, Teofili C, Rondinini C, Fattorini S, Audisio P. 2015. A Red List of Italian Saproxylic Beetles: Taxonomic overview, ecological features and conservation issues (Coleoptera). Fragmenta Entomologica 47:53–126.
- Carpaneto GM, Campanaro A, Hardersen S, Audisio PA, Bologna MA, Roversi PF, Sabbatini Peverieri G, Mason F. 2017. The LIFE Project “Monitoring of insects with public participation” (MIPP): Aims, methods and conclusions. Nature Conservation 20:1–35.
- Castro A, Fernández J. 2016. Tree selection by the endangered beetle Rosalia alpina in a lapsed pollard beech forest. Journal of Insect Conservation 20:201–214.
- Ciach M, Michalcewicz J, Fluda M. 2007. The first report on development of Rosalia alpina (Linnaeus, 1758) (Coleoptera: Cerambycidae) in wood of Ulmus L. in Poland. Polish Journal of Entomology 76:101–105.
- Cizek L, Schlaghamerský J, Bořucký J, Hauck D, Helešic J. 2009. Range expansion of an endangered beetle: Alpine Longhorn Rosalia alpina (Coleoptera: Cerambycidae) spreads to the lowlands of Central Europe. Entomologica Fennica 20:200–206.
- Clement M, Posada D, Crandall KA. 2000. TCS: A computer program to estimate gene genealogies. Molecular Ecology 9:1657–1659.
- Di Santo D, Biscaccianti AB. 2014. Coleotteri saproxilici in Direttiva Habitat del Parco Nazionale del Gran Sasso e Monti della Laga. Bollettino Della Società Entomologica Italiana 146:99–110.
- Don RH, Cox PT, Wainwright BJ, Baker K, Mattick JS. 1991. ‘Touchdown’ PCR to circumvent spurious priming during gene amplification. Nucleic Acids Research 19:4008.
- Drag L, Hauck D, Bérces S, Michalcewicz J, Jelaska LS, Aurenhammer S, Cizek L. 2015. Genetic differentiation of populations of the threatened saproxylic beetle Rosalia longicorn, Rosalia alpina (Coleoptera: Cerambycidae) in Central and South-east Europe. Biological Journal of the Linnean Society 116:911–925.
- Drag L, Hauck D, Pokluda P, Zimmermann K, Cizeck L. 2011. Demography and dispersal ability of a threatened saproxylic beetle. A Mark-Recapture Study of the Rosalia Longicorn (Rosalia alpina). PloS ONE 6:e21345.
- Drag L, JJr Z, Cizek L. 2013. Characterization of nine polymorphic microsatellite loci for a threatened saproxylic beetle Rosalia alpina (Coleoptera: Cerambycidae). Conservation Genetics Resources 5:903–905.
- Duelli P, Wermelinger B. 2005. Rosalia alpina L. Un Cerambicide raro ed emblematico. Sherwood 114:19–25.
- Genovesi P, Angelini P, Bianchi E, Dupré E, Ercole S, Giacanelli V, Ronchi F, Stoch F 2014. Specie e habitat di interesse comunitario in Italia: distribuzione, stato di conservazione e trend. ISPRA, Serie Rapporti, 194/2014.
- Goudet J 2002. Fstat, a Program to estimate and test gene diversities and fixation indices, Version 2.9.3.2. Available: http://www.unil.ch/popgen/softwares/fstat.htm. Accessed Jan 2016 28.
- Hewitt GM. 2000. The genetic legacy of the Quaternary ice ages. Nature 405:907–913.
- Ilić N, Ćurčić S. 2013. The longhorn beetles (Coleoptera: Cerambycidae) of Rtanj mountain: Serbia. Acta Entomologica Serbica 18:69–94.
- Lachat T, Ecker K, Duelli P, Wermelinger B. 2013. Population trends of Rosalia alpina (L.) in Switzerland: A lasting turnaround? Journal of Insect Conservation 17:653–662.
- Librado P, Rozas J. 2009. DnaSP v5: A software for comprehensive analysis of DNA polymorphism data. Bioinformatics 25:1451–1452.
- Lindhe A, Jeppsson T, Ehnström B. 2011. Longhorn beetles in Sweden – Changes in distribution and abundance over the last two hundred years. Entomologisk Tidskrift 131:1–270.
- Magri D. 2008. Patterns of post-glacial spread and the extent of glacial refugia of European beech (Fagus sylvatica). Journal of Biogeography 35:450–463.
- Magri D, Vendramin GG, Comps B, Dupanloup I, Geburek T, Gömöry D, Latalowa M, Litt T, Paule L, Roure JM, Tantau I, Knaap WO, Petit RJ, Beaulieu JL. 2006. A new scenario for the Quaternary history of European beech populations: Paleobotanical evidence and genetic consequences. New Phytologist 171:199–221.
- Mason F, Roversi PF, Audisio P, Bologna MA, Carpaneto GM, Antonini G, Mancini E, Sabbatini Peverieri G, Mosconi F, Solano E, Maurizi E, Maura M, Chiari S, Sabatelli S, Bardiani M, Toni I, Redolfi De Zan L, Rossi de Gasperis S, Tini M, Cini A, Zauli A, Nigro G, Bottacci A, Hardersen S, Campanaro A. 2015. Monitoring of insects with public participation (MIPP; EU LIFE project 11 NAT/IT/000252): Overview on a citizen science initiative and a monitoring programme (Insecta: Coleoptera; Lepidoptera; Orthoptera). Fragmenta Entomologica 47:51–52.
- Michalcewicz J, Ciach M. 2015. Current distribution of the Rosalia longicorn Rosalia alpina (Linnaeus, 1758) (Coleoptera: Cerambycidae) in Poland. Polish Journal of Entomology 84:9–20.
- Müller G. 1950. I coleotteri della Venezia Giulia, II. Coleoptera Phytophaga (Cerambycidae, Chrysomelidae, Bruchidae). Centro Sperimentale Agrario E Forestale, Trieste 4:225–368.
- Oleksa A, Chybicki IJ, Gawroński R, Svensson GP, Burczyk J. 2013. Isolation by distance in saproxylic beetles may increase with niche specialization. Journal of Insect Conservation 17:219–233.
- Peakall R, Smouse PE. 2012. GenAlEx 6.5: Genetic analysis in Excel. Population genetic software for teaching and research-an update. Bioinformatics 28:2537–2539.
- Pilaar Birch SE, Vander Linden M. 2017. A long hard road… Reviewing the evidence for environmental change and population history in the eastern Adriatic and western Balkans during the Late Pleistocene and Early Holocene Quaternary International. DOI: 10.1016/j.quaint.2016.12.035
- Rossi De Gasperis S. 2016. Distribution patterns and population analysis of threatened longhorn beetles in forest habitats of Central Italy. Dissertation, Università degli Studi di Roma Tre, Roma, Italy.
- Rossi de Gasperis S, Carpaneto GM, Nigro G, Antonini G, Chiari S, Cini A, Mancini E, Mason F, Mosconi F, Redolfi De Zan L, Roversi PF, Sabbatini Peverieri G, Solano E, Campanaro A. 2017. Computer-aided photographic identification of Rosalia alpina (Coleoptera: Cerambycidae) applied to a mark-recapture study. Insect Conservation and Diversity 10:54–63.
- Rousset F. 2008. Genepop’007: A complete reimplementation of the Genepop software for Windows and Linux. Molecular Ecology Resources 8:103–106.
- Russo D, Cistrone L, Garonna AP. 2010. Habitat selection by the highly endangered long-horned beetle Rosalia alpina in southern Europe: A multiple spatial scale assessment. Journal of Insect Conservation 15:685–693.
- Sama G. 1988. Coleoptera, Cerambycidae. Catalogo topografico e sinonimico. Fauna d‘Italia, XXVI. Bologna, Italy: Calderini.
- Sama G. 2002. Atlas of the Cerambycidae of Europe and Mediterranean Area: Northern, Western, Central and Eastern Europe. British Isles and Continental Europe from France (excl. Corsica) to Scandinavia and Urals. Zlín, Czech Republic: Kabourek.
- Sama G, Löbl I. 2010. Cerambycidae: Western Palaearctic taxa, eastward to Afghanistan excluding Oman and Yemen and the countries of the former Soviet Union. In: Löbl I, Smetana A, editors. Catalogue of Palaearctic Coleoptera. 6. Chrysomeloidea. Stenstrup, Danmark: Apollo Books. pp. 84–334.
- Shapovalov AM. 2012. Longicorn-beetles (Coleoptera, Cerambycidae) of Orenburg Region: Fauna, distribution, bionomy. Archives of Orenburg Branch of Russian Entomological Society 3:1–224.
- Simon C, Frati F, Beckenbach A, Crespi B, Liu H, Flook P. 1994. Evolution, weighting and phylogenetic utility of mitochondrial gene sequences and a compilation of conserved PCR primer. Annals of the Entomological Society of America 87:651–701.
- Solano E, Mancini E, Ciucci P, Mason F, Audisio P, Antonini G. 2013. The EU protected taxon Morimus funereus Mulsant, 1862 (Coleoptera: Cerambycidae) and its western Palaearctic allies: Systematics and conservation outcomes. Conservation Genetics 14:683–694.
- Speight MCD. 1989. Saproxylic invertebrates and their conservation. Strasbourg, France: Nature and Environment Series - Council of Europe Publishing.
- Staden R, Beal KF, Bonfield JK. 1998. The Staden package. Methods in Molecular Biology 132:115–130.
- Stokland JN, Siitonen J, Jonsson BG. 2012. Biodiversity in Dead Wood. Cambridge, UK: Cambridge University Press.
- Švacha P, Danilevsky ML. 1988. Cerambycoid larvae of Europe and Soviet Union (Coleoptera, Cerambycoidea). Part II. Acta Universitatis Carolinae Biologica 31:121–284.
- Tajima F. 1989. Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics 123:585–595.
- Toonen RJ, Hughes S. 2001. Increased throughput for fragment analysis on ABI prism 377 Automated sequencer using a membrane comb and STRand software. Biotechniques 31:1320–1324.
- van Oosterhout C, Hutchinson WF, Wills DPM, Shipley P. 2004. Micro-checker: Software for identifying and correcting genotyping errors in microsatellite data. Molecular Ecology Notes 4:535–538.
- von Demelt C. 1956. Beobachtungen und Bemerkungen über Rosalia alpina L. Entomologische Blätter 52:170–175.
- Zauli A, Carpaneto GM, Chiari S, Mancini E, Nyabuga FN, Redolfi De Zan L, Romiti F, Sabbani S, Audisio PA, Hedenström E, Bologna MA, Svensson GP. 2016. Assessing the taxonomic status of Osmoderma cristinae (Coleoptera: Scarabaeidae), endemic to Sicily, by genetic, morphological and pheromonal analyses. Journal of Zoological Systematics and Evolutionary Research 54:206–214.
