Abstract
Hamatopeduncularia was erected with Hamatopeduncularis arii as the type species. This genus comprises monogenoidean species mostly found as ectoparasites of marine catfishes belonging to the Ariidae. There is a significant taxonomic ambiguity among Hamatopeduncularia species due to their morphological similarity, but so far only a few morphological studies have succeeded in addressing interspecific variation and relationships. Moreover, little molecular data is available for this genus. A multidisciplinary, integrated study consisting of morphological, morphometric and molecular analyses was conducted on different species of Hamatopeduncularia recovered from the gills of two marine catfishes, Arius jella Day and Plicofollis dussumieri (Valenciennes). Five species of Hamatopeduncularia, two of which represent new species, were investigated: H. arii, H. elongatum, H. thalassini, H. madhaviae sp. nov. and H. bifida sp. nov. Phylogenetic analysis was performed using the 18S rDNA sequence as a molecular marker. The most important results of the present work are: (1) the multidisciplinary description of two novel species; (2) the multidisciplinary redescription of two species and of the type species of the genus; (3) the first molecular characterisation of 18S rDNA sequences of five species of genus Hamatopeduncularia; and (4) molecular support for the monophyly of the genus.
http://zoobank.org/urn:lsid:zoobank.org:act:1333F4CC-E497-4D0A-AD7D-276D44AE6413
http://zoobank.org/urn:lsid:zoobank.org:act:43D18F75-6F4A-4F9B-8C00-6234E5BA6526
Introduction
India, with its vast coastline of 6483 km, hosts approximately 23 species of catfishes belonging to the family Ariidae (Order Siluriformes). Though only 11 species of Ariidae are used in commercial fisheries, they form the major part of the biomass of marine fish landings (Sudarsan et al. Citation1990), therefore representing an important commercial fishery resource along the east coast of India. Fish landings along the Visakhapatnam Coast are mainly concentrated on two species of catfishes, Arius jella Day and Plicofollis dussumieri (Valenciennes), which act as hosts to many metazoan parasites, especially ectoparasites such as monogenoids.
Figure 1. Hamatopeduncularia arii. (A) Composite illustration of entire worm (ventral view). (B) Copulatory organ (ventral view). (C) Vagina. (D–I) Hard parts. (D) Arrangement of anchors and bars (dorsal view): (a) ventral anchor, (b) ventral bar, (c) dorsal anchor, (d) dorsal bar with appendix (adb). (E) Ventral bar. (F) Dorsal bar. (G) Ventral anchors. (H) Dorsal anchors. (I) Appendix on dorsal bar. (J) Marginal hooks.
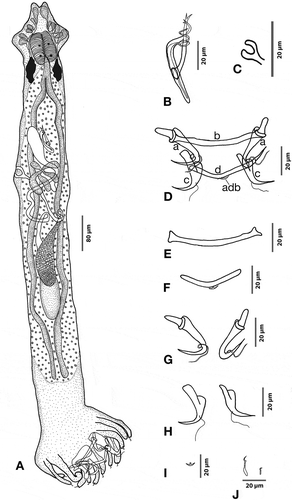
Figure 2. Hamatopeduncularia arii. Scanning electron microscope (SEM) picture to show some features of the parasite posterior end: digitate haptor (dh), armed with a marginal hook (mh), two dorsal anchors (c), dorsal bar (d) with appendix (adb), outer root of anchors (ora). Scale bar: 20 µm.
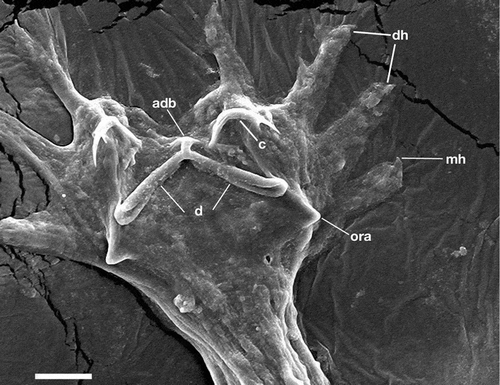
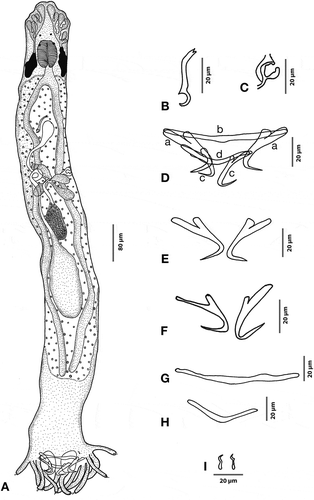
Figure 3. Hamatopeduncularia elongatum. (A) Composite illustration of entire worm (ventral view). (B) Copulatory organ (ventral view). (C) Vagina. (D–I) Hard parts. (D) Arrangement of anchors and bars (dorsal view): (a) ventral anchor, (b) ventral bar, (c) dorsal anchor, (d) dorsal bar. (E) Ventral bar. (F) Dorsal bar. (G) Ventral anchors. (H) Dorsal anchors. (I) Marginal hooks.
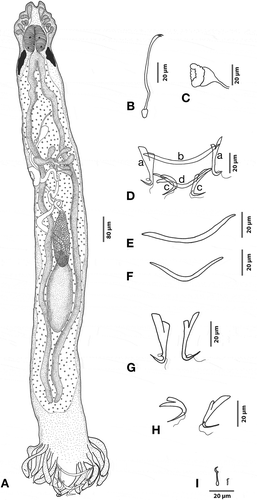
Figure 4. Hamatopeduncularia thalassini. (A) Composite illustration of entire worm (ventral view). (B) Copulatory organ (ventral view). (C) Vagina. (D–I) Hard parts. (D) Arrangement of anchors and bars (ventral view): (a) ventral anchor, (b) ventral bar, (c) dorsal anchor, (d) dorsal bar. (E) Dorsal anchors. (F) Ventral anchors. (G) Dorsal bar. (H) Ventral bar. (I) Marginal hooks.
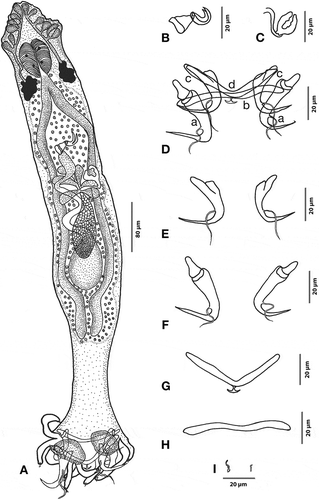
Figure 5. Hamatopeduncularia bifida sp. nov. (A) Composite illustration of entire worm (ventral view). (B) Copulatory organ (ventral view). (C–F) Hard parts. (C) Arrangement of anchors and bars (dorsal view): (a) ventral anchor, (b) ventral bar, (c) dorsal anchor, (d) dorsal bar. (D) Ventral anchors and bar. (E) Dorsal anchors and bar. (F) Marginal hooks. Arrow, gonopore.
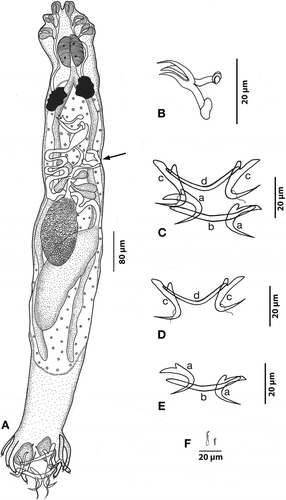
Figure 6. Hamatopeduncularia madhaviae sp. nov. (A) Composite illustration of entire worm (ventral view). (B) Copulatory organ (ventral view). (C) Vagina. (D–I) Hard parts. (D). Arrangement of anchors and bars (ventral view): (a) ventral anchor, (b) ventral bar, (c) dorsal anchor, (d) dorsal bar. (E) Ventral anchors. (F) Dorsal anchors. (G) Ventral bar. (H) Dorsal bar. (I) Marginal hooks.
Among them, Dactylogyridae Bychowsky, 1933 are now attracting significant interest within the scientific community due to their rich biodiversity, with more than 379 species distributed under 31 genera (Mendoza-Palmero et al. Citation2015). Among Dactylogyridae, many new species of Hamatopeduncularia have been described in recent years, but unfortunately most of them were solely based on morphological data (Domingues et al. Citation2016; Yamada et al. Citation2017). However, some descriptions of Dactylogyridae, including both morphological and molecular data, were recently performed (Acosta et al. Citation2017, Citation2018; Francová et al. Citation2017; Verma et al. Citation2017a,Citationb,Citationc; Mendoza-Franco et al. Citation2018).
Hamatopeduncularia was first erected by Yamaguti (Citation1953), with H. arii as the type species. It is a species-rich genus: to date, about 39 species have been described, 24 of them recovered as ectoparasites of fishes belonging to Ariidae. The presence of the haptoral digits, with each digit ending in a marginal hook (simple anchors without any expanded outer roots), is traditionally considered the synapomorphy of the genus. Discrimination among species of Hamatopeduncularia is traditionally performed based on the following morphological characters: male copulatory organ; haptoral components such as anchors, bars and hooks; and vaginal structures (e.g. Domingues et al. Citation2016). Nevertheless, this sometimes has caused serious problems in species discrimination, especially when similar morphological traits were reported (Shinn et al. Citation2010; Strona et al. Citation2013). Thus, in line with the most recent studies on Dactylogyridae (Acosta et al. Citation2017, Citation2018; Verma et al. Citation2017a,Citationb,Citationc; Mendoza-Franco et al. Citation2018), a multidisciplinary characterisation of the genus including the study of molecular markers for species description is recommended.
Ribosomal RNA genes are generally considered useful markers to study molecular taxonomy and phylogeny of both prokaryotic (e.g. Ferrantini et al. Citation2009; Castelli et al. Citation2018a,Citationb; Chiellini et al. Citation2019; Fokin et al. Citation2019; Lanzoni et al. Citation2019) and eukaryotic organisms (e.g. Modeo et al. Citation2013a,Citationb; Nitla et al. Citation2018; Fokin et al. Citation2019), parasitic metazoans included (e.g. Blair & Barker Citation1993; Zhu et al. Citation1998; Caffara et al. Citation2016; Sailaja et al. Citation2016; Mendoza-Franco et al. Citation2018; Tedesco et al. Citation2018). As for monogenoidean parasites, however, there are no published molecular studies on the genus Hamatopeduncularia except for the paper by Mendoza-Franco et al. (Citation2018) which provides partial sequences of the 28S rRNA gene from monogeneans of ariid, sparid and haemulid hosts to explore their systematic position within the Monogenea.
In the present study, we recovered different morphotypes of these parasites from the marine catfishes A. jella and P. dussumieri caught during a 2-year period of investigation (January 2013–December 2014). To validate the taxonomic status of these Hamatopeduncularia parasites, we applied an integrative taxonomic investigation combining morphological–morphometric with molecular study, i.e. performing morphological observations, also with scanning electron microscope (SEM), processing morphometric data with multivariate analysis methods, and, for the first time on this genus, sequencing the 18S rDNA and producing a molecular phylogenesis on this basis. Morphometric data were also used for the quantitative characters, and multivariate analysis methods were applied to identify the species-specific pattern of traits for each investigated species.
A complete morphological account, supported by statistical evidence, was obtained for two novel Hamatopeduncularia species, H. madhaviae sp. nov. and H. bifida sp. nov., and for three already known species of the genus, H. arii, H. elongatum Lim, Citation1996 and H. thalassini Bychowsky and Nagibina, Citation1969. Results were compared with previous morphological works (Bychowsky & Nagibina Citation1969; Lim Citation1996; Yao et al. Citation1998; Domingues et al. Citation2016). The molecular characterisation that was performed supported the morphological results in most cases.
Material and methods
Preparation for morphological characterisation
Freshly caught ariid fishes were brought to the laboratory where they were identified and examined for the presence of monogenoidean parasites. For the collection of monogenoidean worms, gill arches were removed and placed in a petri dish containing 8‰ saline water to avoid salinity stress, which may lead to contraction of forms. This saline solution was prepared by mixing one part seawater (salinity: 32‰) with three parts tap water. Sometimes an 8‰ solution of NaCl in water was also used (Cribb & Bray Citation2010). All gill parts were scraped with a needle to detach the parasites. The contents were transferred to an evaporating bowl and washed well by the process of decantation. This step was repeated until the contents appeared clear and the parasites were cleaned of gill mucus. Then the contents were taken in a small Petri dish and examined under a binocular stereo microscope (Nikon SMZ800) at 5× magnification. The detached monogenoids were picked up using a fine needle and transferred individually into a drop of glycerine on a slide for the preparation of semi-permanent material. The preparation was then covered with a coverslip and sealed for the examination of sclerotised structures. Some parasites mounted on slides with water droplet were also observed in vivo under a histological microscope (Nikon YS100) at 10–100× magnification. They were stained with alum carmine, dehydrated through ascending grades of alcohol and mounted in Canada balsam for preparation of permanent slides.
From these permanent slides schematic line drawings were made with a camera lucida connected to a microscope Nikon ECLIPSE 50i. Measurements were taken with the use of an ocular micrometre; a maximum number of 10 specimens for each species of parasite were used for this purpose. All measurements are given in micrometres (μm) and presented as the average, followed in parentheses by the range. Body length includes haptor. Measuring method and terminology concerning the hard structures of the haptor (i.e. anchor, marginal hooks, and dorsal bar) are according to Gusev (Citation1977). Identification of the parasites was performed by studying forms in freshly dead and, sometimes, live condition and by observing details from mounted specimens under the light microscope. The utmost care was taken to study the observed modifications of haptoral armature and of copulatory organs, which play an important role in the identification of monogenoidean parasites. Papers by Yamaguti (Citation1953), Tripathi (Citation1959), Gusev (Citation1976), Kearn and Whittington (Citation1994) and Lim (Citation1996) were considered both for species identification/description and establishment of novel taxa.
Good-quality permanent slides with representatives of each species of parasite have been deposited in the helminthological collections of the Central Zone Regional Centre, Zoological Survey of India, 68–169, Vijoy Nagar, Jabalpur – 482 002, Madhya Pradesh; the assigned museum registration numbers are listed in the Results section.
Morphometric analysis
To investigate differences and relationships among the five species of Hamatopeduncularia, we collected morphometric data for 53 characters, including details of general morphology, copulatory structures and haptoral hard parts, from 10 specimens for each species. These data were subsequently analysed with multivariate statistics such as Euclidean cluster analysis, multidimensional scaling (MDS) and principal component analysis (PCA). These analyses were performed using PRIMER v. 6 (Plymouth Routines in Multivariate Ecological Research) and STATISTICA v. 8 software.
The dendrogram clustering method uses the similarities or distances between the species taken as samples while forming the clusters. Similarities are a set of rules that serve as criteria for grouping or separating the species, based on multiple dimensions. The most straightforward way of computing distances between the objects in a multidimensional space is to compute Euclidean distances. A correlation matrix (morphological measurements vs. parasite species) was prepared based on obtained data, and an overall mean was derived for each parameter. The data were then analysed by hierarchical clustering through group average linking, following Euclidean dendrogram cluster analysis procedures implemented in PRIMER v. 6. The output of this analysis is shown as a dendrogram of relationships between groups/species. Ordination plots (MDS) were then produced from these similarity measures through PRIMER dendrogram protocol. Significance tests of samples were made using the analysis of similarity (ANOSIM) randomisation test (Clarke & Green Citation1988).
PCA is a statistical ordination technique that reduces the number of dimensions (variables) in a set of data, finds linear combinations of the more correlated variables that explain most of the variance, and eventually assigns a component score to each original individual (Foottit & Sorensen Citation1992). The morphometric data were subjected to PCA using the software STATISTICA v. 8. PCA was used in estimating the morphometric variation among species and to identify the variables that substantially contribute to this variation. The results are presented in the form of graphs and tables. Finally, a PCA plot was obtained using the components showing high variance.
SEM preparation of Hamatopeduncularia arii
Freshly collected H. arii samples were used to obtain SEM preparations according to the protocol by Fisher et al. (Citation2012) with slight modifications. Parasites were washed thoroughly in 8‰ saline water until all the debris was removed, fixed in 2.5% glutaraldehyde for 2 hours at 4°C, washed with 0.1 M phosphate buffer (3 changes of 15 min each at 4°C), and dehydrated in an ascending series of acetone. Then specimens were dried in a critical point drying apparatus, mounted on aluminium stubs, and gold coated. The prepared specimens were placed in a JSM-6610LV SEM for observation.
Molecular and phylogenetic analysis
After the collection of parasites from the gill filaments, each specimen to be processed for molecular analysis was observed and identified under a Nikon YS100 stereo zoom microscope, and then preserved in 95% ethanol at –20°C. DNA was extracted using a NucleoSpin™ PlantII DNA extraction kit (Macherey-Nagel GmbH & Co., Düren NRW, Germany) according to the manufacturer’s instructions, and stored at –20°C until further processing. The 18S rDNA was amplified according to Modeo et al. (Citation2013b) using the following primers: Forward 18S F9 as forward primer (5ʹ – CTGGTTGATCCTGCCAG – 3ʹ) (Medlin et al. Citation1988) and 18S R1513 Hypo as reverse primer (5ʹ – TGATCC TTCYGCAGGTTC – 3ʹ) (Petroni et al. Citation2002).
The polymerase chain reaction (PCR) cycle, performed according to the manufacturer’s instructions (Takara Bio, USA), was set up in a Gene Amp system C1000™ Thermal Cycler (Bio-Rad, Hercules, CA) with the following steps: 3 min at 94°C for the initial denaturation, followed by 35 cycles of 30 sec at 94°C for denaturation, 30 sec at 55°C for primer annealing, 2 min at 72°C for extension, and a final extension for 6 min at 72°C. An aliquot (5 μL) of each amplicon was checked on 1% agarose gel. Gel electrophoresis run was performed for 40 min at 150 V along with a 100–10,000-bp DNA molecular weight marker and, later, stained with ethidium bromide for 30 min. PCR bands were visualised under a UV light transilluminator and photographed with a Gel Doc system. PCR product was purified using a Euro Gold Cycle Pure Kit (Euroclone SpA, Italy) and stored at –20°C until it was sent for sequencing.
Semi-nested PCR was performed using the primer combinations 18S F9 + 18S R1052 and F300 + 18S R1513 Hypo for the PCR samples of H. elongatum and H. bifida sp. nov. to obtain more abundant amplification products of the target region [18S R1052: 5ʹ – AACTAAGAACGGCCATGCA – 3ʹ – Rosati et al. (Citation2004); F300: 5ʹ – AGGGTTCGATTCCGGAGA – 3ʹ – Andreoli et al. (Citation2009)]. PCR cycles were set as previously, except for the extension time: 1.5 min. PCR products were sequenced with the GATC Biotech sequencing services, using three internal primers as in Nitla et al. (Citation2018): 18S R536 (5ʹ – CTGGAATTACCGCGGCTG – 3ʹ), 18S F783 (5ʹ – GACGATCAGATACCGTC – 3ʹ), and 18S R1052.
The resulting 18S rDNA electropherograms were checked and assembled using Chromas Lite 2.1 software. The sequences thus obtained were deposited in the National Center for Biotechnology Information (NCBI) GenBank database (for the accession numbers see ). Hamatopeduncularia sequences from the present study and the latest sequences available in the NCBI database belonging to the subclass Polyonchoinea were automatically aligned with the ARB software package (Ludwig et al. Citation2004) against the SILVA 102 SSU rRNA database (Pruesse et al. Citation2007). Then, alignment of monogenoidean sequences was manually refined. For phylogenetic analyses 85 sequences were employed: we selected 55 sequences belonging to the order Dactylogyridea, the present five sequences, plus 25 more as outgroup (23 Gyrodactylidea plus two incertae sedis). The alignment was reduced in length according to the shortest sequence, producing an 1806-character matrix. Maximum likelihood (ML) analyses (PHYML 5.3.2; Guindon & Gascuel Citation2003) and Bayesian inference (BI) analyses (MrBayes 3.2; Ronquist et al. Citation2012) were performed, with the GTR + I + G substitution model, as indicated by the Akaike information criterion (AIC), calculated by jModelTest 2.2 (Darriba et al. Citation2012). ML analysis was performed with 1000 pseudoreplicates, while for BI analysis, three different Markov chain Monte Carlo runs were used, with one cold chain and three heated chains each, running for 1,000,000 generations. The ARB NJ algorithm (Ludwig et al. Citation2004) with the “similarity” correction was employed to calculate the similarity matrix.
Table I. 18S rDNA sequences of studied Hamatopeduncularia species.
Results
The description/redescription of the five species of parasites recovered on the marine catfishes A. jella and P. dussumieri from the Visakhapatnam Coast, Bay of Bengal, between January 2013 and December 2014 follows. For each parasite species, a detailed account, the indication of the host from which it was isolated, some ecological notes (prevalence of infection and mean intensity), and some remarks concerning previous descriptions are provided. All measurements are in µm.
Class Monogenoidea Bychowsky, Citation1937
Subclass Polyonchoinea Bychowsky, Citation1937
Order Dactylogyridea Bychowsky, Citation1937
Dactylogyridae Bychowsky, 1933
Hamatopeduncularia Yamaguti, Citation1953
Hamatopeduncularia arii Yamaguti, Citation1953
( and ; )
Table II. Morphometric data for five species of Hamatopeduncularia (present study). Measurements are reported in micrometres. For each species 10 specimens were analysed.
Table II. (continued)
Table II. (continued)
Host
Arius jella.
Site of infection
Gill lamellae.
Locality
Off the Visakhapatnam Coast, Bay of Bengal, Andhra Pradesh, India (17°47ʹN, 83°50ʹE).
Voucher material
Slide A-18,080 with 10 specimens.
Prevalence of infection
58 of 835 (7%).
Mean intensity
Eighteen parasites per infected host (total number of recovered H. arii specimens: 1063).
18S rDNA sequence
NCBI GenBank accession number KT252895.
Redescription
Based on 10 adult specimens. Fusiform parasite measuring 1342 (880–1920) in length and 140 (96–176) in width. Prohaptor with three bilateral pairs of head organs; cephalic glands lying posterior to pharynx. Two pairs of eyespots situated at a distance of 69 (56–80) from the anterior end. Mouth subterminal, well-developed pharynx measuring 68 (52–84) in length and 50 (36–100) in width. Oesophagus short, leads into simple intestinal crura, which blindly terminate prior to the haptor at peduncular region. Gonads overlapping. Testis single, elongate, 190 (120–200) in length and 56 (32–88) in width, in post-equatorial position. Vas deferens long, arising from the anterior margin of the testis, running outside around the left caecum. Seminal vesicle dextral to copulatory complex. Copulatory tube 50 (40–60) in length and 6 in width, provided with an elongated accessory piece, measuring 45 (36–64) in length and 8 (6–8) in width. Accessory piece slightly coiled at the distal end of the copulatory tube. Ovary pretesticular, slightly overlapping, the anterior margin of the testis measuring 98 (60–128) in length and 36 (28–48) in width. Mehlis gland, seminal receptacle and transverse vitelline duct joining in front of the ovary. Genital atrium occupying the area just below the caecal bifurcation. Vagina 46 (28–60) in length and 26 (20–32) in width. Follicular vitellaria occupying the lateral margins of the intestinal crura, being confluent behind testis. Haptor 172.4 (144–192) long, 308 (192–420) wide, digitate, provided with 12 short finger-like projections known as digits (Lim Citation1996); 14 marginal hooklets, 12 of which are located on haptoral digits and one pair located near ventral anchors. A short peduncle present between proper body and haptor. The two ventral anchors are supported by a simple ventral bar having a length of 7 (5–8) and a maximum width of 77 (72–96). Ring pads found on the inner roots of the ventral anchors. A pair of dorsal anchors also present, with inner length 47 (36–52), inner root length 20 (12–24), outer root length 12 (8–16), and a point 25 (24–28) long. Bar supporting the dorsal anchors with a length of 7 (5–8) and a width of 75 (72–96). Dorsal bar with anchor-shaped appendix. The haptoral hard parts of H. arii were also examined under SEM ().
Remarks
The present form has been identified as H. arii based on the structure of the copulatory complex, position of gonads, and structure and armature of haptor, which are very similar to those previously reported (Yamaguti Citation1953; Bychowsky & Nagibina Citation1969; Paperna Citation1977; Lim Citation1996). In the specimens here investigated, both ring pads and appendix on dorsal bar were observed (see Discussion).
Hamatopeduncularia elongatum Lim, Citation1996
(; )
Host
Arius jella.
Site of infection
Gill lamellae.
Locality
Off the Visakhapatnam Coast, Bay of Bengal, Andhra Pradesh, India (17°47ʹN, 83°50ʹE).
Voucher material
Slides A-18,078, A-18,079.
Prevalence of infection
261 of 835 (31%).
Mean intensity
Seven parasites per infected host (total number of recovered H. elongatum specimens: 1935).
18S rDNA sequence
NCBI GenBank accession number MK084780.
Redescription
Based on 10 adult specimens. Body slender, elongate with a length of 1010 (880–1220) and a width of 116 (80–120). Prohaptor with three bilateral pairs of head organs; cephalic glands lying posterior to pharynx. Two pairs of prominent eyespots at a distance of 51 (32–64) from anterior end. Mouth subterminal, pharynx large, circular and divided into two semi-circular halves 62 (48–84) in length and 40 (36–54) in width. Oesophagus short, leading into simple intestinal caeca, blindly terminating at peduncular region. Gonads intercaecal; testis elongate, large, post equatorial, measuring 340 (220–384) in length and 56 (32–72) in width. Vas deferens arising from the anterior region of the testis and looping around the left caecum. Vagina cup-like and muscular, dextrally opening. Vaginal tube slightly sclerotised. Long copulatory tube 70 (58–98) in length, slightly anteriorly bent, of “elegans-type” according to Oliver (Citation1987). A pair of small cement glands associated with ventral anchors. Pretesticular, oval ovary, 90 (60–120) in length and 41 (24–56) in width. Vitellaria co-extensive with intestinal crura, from the pharyngeal to the peduncular region. Haptor well developed, 252.6 (160–320) long and 358.8 (320–480) wide, with a short peduncle, and digitate with 12 stout, stumpy and bulbous projections, each measuring 253 (160–320) in length and 359 (320–480) in width. Seven pairs of marginal hooks, six pairs located on peduncles and one pair near the ventral anchors. Anchors in two pairs, ventral anchor showing an inner length of 43 (36–48) µm. Inner root length 16 (12–24); outer root length 12 (12–12), supported by a simple ventral bar 8 (6–8) in length and 69 (60–80) in width. Dorsal anchors 40 (36–44) in length, inner root 13 (12–16) in length, outer root 9 (8–12) in length, point 18 (12–20) long. Dorsal anchors supported by a dorsal bar showing a length of 8 (6–8) and a width of 68 (52–84).
Remarks
The present form was identified as H. elongatum based on the similarities of copulatory tube and haptor armature proposed by Lim (Citation1996) (see Discussion).
Hamatopeduncularia thalassini Bychowsky and Nagibina, Citation1969
(; )
Host
Arius jella.
Site of infection
Gill lamellae.
Locality
Off the Visakhapatnam Coast, Bay of Bengal, Andhra Pradesh, India (17°47ʹN, 83°50ʹE).
Voucher material
Slides A-18,072, A-18,073, A-18,074, A-18,075.
Prevalence of infection
316 of 835 (38%).
Mean intensity
Twelve parasites per infected host (total number of recovered H. thalassini specimens: 3739).
18S rDNA sequence
NCBI GenBank accession number KT252901.
Redescription
Based on 10 adult specimens. Body elongated, 1010 (1280–848) in length and 127 (160–112) in width. Prohaptor with three bilateral pairs of head organs; cephalic glands lying posterior to pharynx. Two pairs of eyespots situated at a distance of 35 (48–24) from the anterior region, anterior pair smaller than posterior. Mouth sub-terminal. Pharynx globular, 56 (48–68) in length and 46 (40–56) in width. Oesophagus short, 39 (28–16) in length, leads into simple, intestinal crura blindly terminating prior to the haptor at the beginning of peduncular region. Testis large, elliptical measuring 106 (88–128) in length and 58 (44–72) in width. Vas deferens running on right side, forming a loop and turning medially. Pre-testicular, small and compact ovary, 87 (80–108) in length and 42 (40–60) in width. Vitelline follicles not extending into the haptor. Mehlis gland, seminal receptacle, and transverse vitelline duct joining in front of the ovary; uterus opening into a common genital atrium. Vagina 36 (28–48) long and 26 (16–36) wide, and dextral in position. Short copulatory tube measuring 40 (32–44) in length and 14 (8–16) in width, with spatula-like accessory piece 22 (20–28) in length and 15 (12–24) in width. Haptor 163 (116–220) long and 222 (200–280) wide, digitate, with six pairs of long haptoral digits each ending in a hook measuring 20 (16–24) in length; two pairs of anchors plus one pair of cement glands. Dorsal anchor measuring 64 (60–68) in length with long inner root 26 (24–28) in length, slightly expanded outer root 12 (10–16) in length and long narrow recurved point 28 (12–32) long. Ventral anchor measuring 48 (48–52) in length; with inner root 26 (24–28) long, having ring pads, very short outer root 10 (8–12) in length and wide recurved point, 29 (24–32) long. “V”-shaped dorsal bar with expanded wings, 7 (4–8) in length and 89 (76–100) in width. Anchor-shaped appendix, 13 (12–20) in length and 6 (6) in width. Ventral bar straight, with bulged ends, 8 (8) in length and 80 (84–76) in width, 14 marginal hooklets, 12 located on haptoral digits and one pair located near ventral anchors.
Remarks
This parasite was identified as H. thalassini mainly based on size of anchors, presence of specialised haptoral structures such as ring pads, appendix, shape of the copulatory organ, accessory piece with spatulated end, and vaginal armament, which resemble the corresponding characters of the previous description by Bychowsky and Nagibina (Citation1969). The present study constitutes both the first report of this species from the catfish A. jella and the first report from Visakhapatnam coast, Bay of Bengal, India (see Discussion).
Hamatopeduncularia bifida sp. nov.
(; )
Type host
Arius jella.
Site of infection
Gill lamellae.
Type locality
Off the Visakhapatnam Coast, Bay of Bengal, Andhra Pradesh, India (17°47ʹN, 83°50ʹE).
Type material
Holotype, slide W10423/1.
Prevalence of infection
213 of 835 (25.5%).
Mean intensity
Ten parasites per infected host (total number of recovered H. bifida sp. nov. specimens: 2204).
18S rDNA sequence
NCBI GenBank accession number MK084781.
Etymology
The term “bifida” (N.L. fem. adj., meaning “bifurcated”) refers to the bifurcated tip of the accessory piece of the male copulatory organ.
Description
Based on 10 adult specimens. Body elongated, measuring 1891 (1552–3040) in length and 220 (112–288) in width. Haptor fairly set off from the body by a haptoral peduncle in the posterior region. Prohaptor lodged with three bilateral pairs of head organs; cephalic glands lying very posterior to pharynx. Two pairs of eyespots. Each head gland provided with a separate duct extending posteriorly. Pharynx spherical, muscular, bifurcate, 92 (64–120) in length and 76 (48–108) in width. Caeca 1394 (1120–1760) in length, blindly ending at peduncle level. Oval, inter-caecal, equatorial testis, 334 (240–520) in length and 103 (44–128) in width. Copulatory tube 60 (40–76) in length and 8 (8–10) in width, with an accessory piece 54 (40–60) long and 9 (6–10) wide. Accessory piece showing bifurcation at the tip and appearing slightly tilted at right angle; the base is bubble like. Pre-equatorial, oval ovary, 143 (92–200) in length and 47 (32–68) in width. Vagina 46 (32–60) in length and 41 (32–56) in width, dextral in position and muscular. Oval seminal receptacle located at ovary level. Vitelline follicles co-extensive with caeca. Haptor 281 (112–440) long and 342 (148–560) wide, with six pairs of haptoral digits. Armature of haptor consisting of two pairs of anchors, double transverse bars, and marginal hooklets. Each dorsal anchor measuring 47 (44–48) in length, with inner root 26 (20–28) long, having spatulated ends; slightly differentiated outer root measuring 16 (8–24) in length; narrow, recurved point, 23 (20–24) long. In the shaft region, each dorsal anchor equipped with well-developed sleeve sclerite. Dorsal transverse bar 8 (8) in length and 75 (68–80) in width, slightly “V” shaped. Each ventral anchor measuring 47 (44–48) in length, with inner root 26 (20–36) in length, outer root 16 (12–24) in length, recurved point 23 (20–24) long. In the shaft region, each ventral anchor is protruded and equipped with sleeve sclerite. Rod-like ventral transverse bar 8 (8–10) in length and 64 (56–72) in width. Each haptoral digit ending with a hook. Seven pairs of marginal hooklets. One pair of larval hooks near the ventral anchors. One pair of cement glands associated with ventral anchors.
Remarks
This parasite is characterised by a copulatory organ provided with a tubular accessory piece with a bifurcated tip, slightly tilted at a right angle, and a broad base. The haptoral armature consists of six pairs of thin, long peduncles, each carrying a marginal hook, a “V”-shaped dorsal transverse bar and a pair of cement glands. It further differs in the structure of the accessory piece, which is bifid and not reported in other species of Hamatopeduncularia so far. Additionally, the length of the peduncle connecting the proper body with haptor is considerably long in this species. Although a certain grade of variation is present in haptoral armature ( and ), based on the differences with other species of Hamatopeduncularia the present morphospecies is considered new and designated as H. bifida (see Discussion).
Hamatopeduncularia madhaviae sp. nov.
(; )
Type host
Plicofollis dussumieri.
Site of infection
Gill lamellae.
Type locality
Off the Visakhapatnam Coast, Bay of Bengal, Andhra Pradesh, India (17°47ʹN, 83°50ʹE).
Type material
Holotype, slide W10425/1.
Prevalence of infection
59 of 138 (43%).
Mean intensity
Thirteen parasites per infected host (total number of recovered H. madhaviae sp. nov. specimens: 752).
18S rDNA sequence
NCBI GenBank accession number KT252898.
Etymology
The term “madhaviae” is in honour of Professor Rokkam Madhavi, in recognition of her contribution to helminthology.
Description
Based on 10 adult specimens. Body elongate 1776 (1040–3040) in length and 178 (128–272) in width. Prohaptor with three bilateral pairs of head organs; cephalic glands lying lateral to pharynx. Eyespots in two pairs, posterior larger. Pharynx oval, 86 (56–112) in length and 58 (40–72) in width, muscular. Intestinal caeca simple and straight. Pre-testicular, pre-equatorial, inter-caecal, oval ovary, measuring 118 (80–136) in length and 53 (24–88) in width. Dextral vagina, 40 (32–48) long and 30 (20–40) wide, opening to exterior through a small cup-shaped opening. Testis 226 (152–320) long and 80 (62–120) wide; post-equatorial, postovarian, and club-shaped. Dextral, inter-caecal vas deferens, dilating to form club-shaped seminal vesicle. Copulatory tube measuring 55 (40–68) in length and 9 (8–10) in width, in the form of a double-walled chitinoid structure with a hook-like base. Accessory piece absent. Follicular vitellaria extending from behind pharynx up to peduncle. Haptor 198 (140–280) long and 293 (148–400) wide, digitate, with six pairs of haptoral peduncles, two pairs of boreal type dissimilar and unequal anchors, and dorsal plus ventral transverse bars. Dorsal anchor 43 (40–48) long, with wide base. Short, flattened outer root 11 (8–12) in length and long, rounded inner root 20 (16–20) in length; base 11 (8–12) in length, narrowing into a curved shaft, with long tapering point 23 (20–24) long. Ventral anchor 47 (44–48) in length, base divided into a slightly broad, outstretched inner root measuring 23 (20–24) in length, a short outer root 10 (8–12) in length, and a recurved point 24 µm long. Straight but slightly bent in the middle dorsal bar, measuring 8 (6–8) in length and 94 (80–112) in width. Ventral bar 10 (8–10) in length and 83 (76–88) in width, curved like a boomerang. Seven pairs of marginal hooks: six pairs shifted posteriorly towards edge of haptoral peduncles, and the last pair lying near the dorsal anchor shafts. Each hook composed of an inconspicuous oval base, a relatively median-sized solid shaft, and a sickle-shaped termination.
Remarks
This parasite species considerably differs from all other species of the genus reported so far in the structure of haptoral armature and copulatory organs; thus, we are proposing it as the new species H. madhaviae (see Discussion).
Morphometric analysis
Euclidean cluster analysis
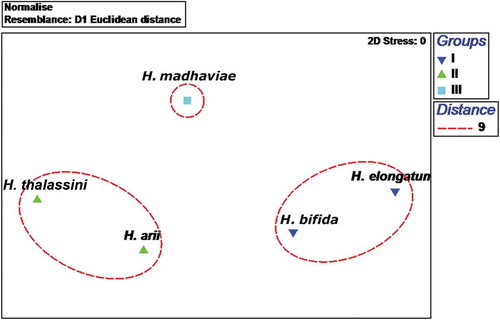
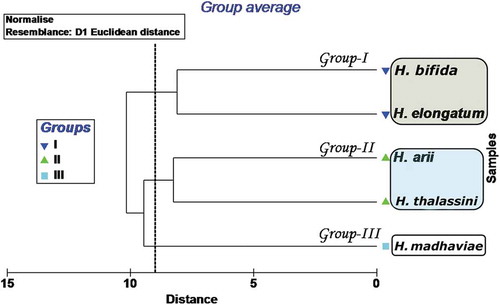
The Euclidean cluster analysis identified three clusters at a distance of 9 with stress at 0, which were separated clearly with ANOSIM: Global R = 1.0; P = 0.1%. The first cluster (Group I) was represented by H. elongatum and H. bifida sp. nov.; the second cluster (Group II) was an assemblage of two species, H. thalassini and H. arii; and the third cluster (Group III) included only one species, H. madhaviae sp. nov. (). The plot obtained as result of multidimensional scaling supports the Euclidean cluster analysis (). Hamatopeduncularia elongatum and H. bifida sp. nov. in Group I showed similarities in the structure of their haptoral hard parts, with simple bars and anchors. In Group II, H. arii and H. thalassini possessed similar haptoral armature (i.e. bars with expanded wings, anchors with ring pads on the inner root of ventral anchors, and anchor-shaped appendix). Hamatopeduncularia madhaviae sp. nov., forming the separate cluster Group III, was characterised by a long dorsal bar, a curved ventral bar, a simple, short copulatory tube, and a narrow peduncle.
Figure 7. Euclidian cluster analysis: dendrogram of relationships between groups. Three clusters were identified at a distance of 9 with stress at 0: Group I, represented by Hamatopeduncularia elongatum and H. bifida sp. nov.; Group II comprising H. thalassini and H. arii; Group III, represented by H. madhaviae sp. nov.
Principal component analysis (PCA)
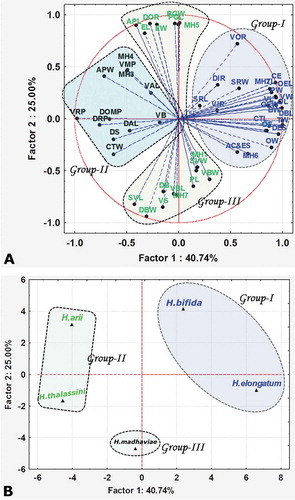
In Euclidean cluster analysis, the discrimination of species is mainly based on their morphometric characters presented in a two-dimensional model with fairly convincing groups in the form of a Euclidean dendrogram and mean dimensional scaling. However, PCA is more commonly used to understand morphological variations at each parameter level. Based on the morphometric variations of the data, principal components were identified in the PCA. Those having an eigenvalue > 1 were utilised to describe the data. The correlation values for each parameter tested with each axis and their discrimination into principal components (PC1, PC2, PC3) are provided in and . Group I (eigenvalue 21.6 and total variance 40.7%) isolated the species H. elongatum and H. bifida sp. nov. from other species based on the following traits: body length and width, distance between anterior cleft and eyespots, distance between eyespots, pharynx width, oesophagus, caeca, testis length and width, ovary length and width, seminal receptacle length and width, vagina length and width, copulatory tube length, haptor length and width, dorsal bar length, dorsal and ventral inner root, ventral outer root, 2nd and 6th pairs of marginal hooks. Group II (eigenvalue 13.3 with a variance of 25%) corresponded to the species H. thalassini and H. arii. Principal components supporting this group were pharynx length, seminal vesicle length and width, egg length and width, prostate gland length and width, dorsal and ventral bar length, ventral bar width, dorsal anchor length, dorsal base, ventral shaft, 1st, 5th and 7th pairs of marginal hooks. Hamatopeduncularia madhaviae sp. nov. constituted a separate cluster, Group III, with an eigenvalue of 11.9 and variance of 22.4%. Principal components that supported this group were copulatory tube width, accessory piece length and width, dorsal outer root, dorsal main piece, dorsal shaft, dorsal recurved point, ventral anchor length, ventral main piece, ventral base, ventral recurved point, 3rd and 4th pairs of marginal hooks. The total variance for the first three axes together scored ~88.2%. The morphological characters responsible for the discrimination of the three groups in the cluster analysis were identified in the PCA. Hence, ordination analysis is recognised as the best tool to explain the projection of individual variable scores and represented in the form of variance (%).
Table III. Axis value results of the principal component analysis (PCA).
The total eigenvalues for each principal component are shown in . The results show the highest total value of 21.6 for PC1, and eigenvalues for PC2 and PC3 of 13.3 and 11.9, respectively. The total variance for the first three axes together scores ~88.2%, and there is another axis with an eigenvalue of 6.3 with a total variance of 11.8. Compared with the other three axes, the values obtained for the fourth axis are low. Hence, this axis was merged with the third axis in this analysis (). The results obtained from the PCA are in close association with those from the cluster analysis, and the morphological characters which are responsible for the discrimination of three groups in the cluster analysis were identified in the PCA, as major parameters that support the differentiation of the species from one another. The scores of individual morphological characters obtained through the PCA are shown in . The score plot revealed clear separation among the species based on morphometric characters (). The scatter diagram of PCA 1 versus PCA 2 shows the relationships among the species ().
Table IV. Summarised data of the principal component analysis (PCA).
Table V. Principal component values for Hamatopeduncularia species discrimination.
Molecular and phylogenetic analyses
The 18S rDNA sequences characterised from the studied species of Hamatopeduncularia shared similarity values higher than 96.4% (). In our phylogenetic analysis (), BI and ML topologies were coherent except for: (1) the clade with Euryhaliotrema grandis, E. microphallus and E. berenguelae; (2) the clade with Bravohollisia maculatus, B. tecta, B. plectorhynchus, Haliotrema fleti, H. cromileptis, H. macasariensis, Lethrinitrema zhanjiangense and Pseudohaliotrema sphincteroporus; and (3) the clade with Mizelleus indicus and M. longicirrus. In the first and the second case, BI analysis showed a polytomy; in the third case, the Mizelleus clade represented the sister clade of Hamatopeduncularia. Nevertheless, statistical values were low in each analysis, showing that those phylogenetic associations were weakly supported in both BI and ML topologies. All available Hamatopeduncularia sequences clustered together, forming a monophyletic, well-supported clade (1.00/100) inside the family Dactylogyridae. The sequences of H. elongatum and H. bifida sp. nov. were almost identical, with only one nucleotide mismatch. In the present analysis, phylogenetic associations among Hamatopeduncularia species were stable and supported by statistical value: H. elongatum and H. bifida sp. nov. clustered together as sister group to the novel species H. madhaviae sp. nov., which, in turn, together with H. elongatum and H. bifida sp. nov., resulted as sister group to the clade represented by H. arii and H. thalassini.
Figure 10. Phylogenetic tree of the order Dactylogyridea based on 18S rDNA sequences. Numbers associated with nodes represent posterior probabilities and maximum likelihood bootstrap values, respectively (only values above 0.80–75 are shown). Sequences obtained in the present work are in bold.
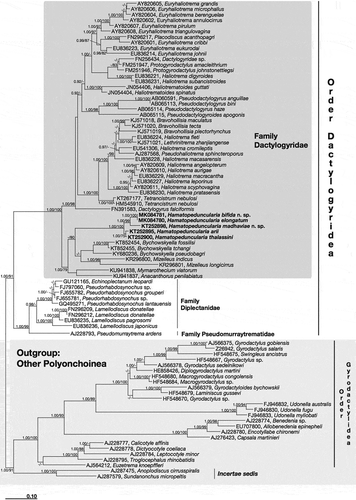
Table VI. Similarity matrix of 18SrDNA sequences of studied species of Hamatopeduncularia and their closest relatives. Similarity values are calculated using the ARB NJ algorithm; sequences obtained in the present work are in bold.
The sister genus of Hamatopeduncularia was not properly resolved in our analysis at the molecular level, being represented either by genus Bychowskyella or by genus Mizelleus, with genera Mymarothecium and Anacanthorus as weaker options; however, the family Dactylogyridae comprises more than 200 genera and our analysis unfortunately could include only a small fraction of them, i.e. those for which the molecular marker sequence is available. Identity values between the 18S rDNA sequences of the five Hamatopeduncularia species herein studied and those of their closest relatives according to the present molecular investigation ranged between 86.4% and 90.5% (). Hamatopeduncularia, Bychowskyella and Mizelleus, together with Mymarothecium and Anacanthorus, formed a monophyletic clade sister to all other sequenced members of the family Dactylogyridae.
Discussion
Considerations on the investigated Hamatopeduncularia species
In the present study, with a sampling campaign covering a 2-year period, the catfish P. dussumieri was found to be infected with a single Hamatopeduncularia species, H. madhaviae sp. nov., whereas the catfish A. jella hosted four species, H. arii, H. elongatum, H. thalassini and H. bifida sp. nov. Hence, according to our research, catfishes of the species A. jella can host a larger number of Hamatopeduncularia species then P. dussumieri.
However, the susceptibility of fish hosts to parasites can be influenced by various factors such as host sex, ontogenetic alterations in behaviour, physiology and ecology (Takemoto et al. Citation1996), and the evolutionary history of the host–parasite relationship is even more important.
As for the prevalence and mean intensity of infection for the different investigated monogenoids, H. madhaviae shows the highest prevalence value (43%) and H. arii shows the highest mean intensity value (18) with respect to the other investigated species.
Species identification of monogenoidean parasites is traditionally based only on morphological characters, including the haptoral armature and copulatory organ, which are considered taxonomic markers (Gerasev Citation1992; Lim Citation1995, Citation1996). In the present study, the five species of Hamatopeduncularia were investigated through a multidisciplinary integrated study approach: morphological analysis was further supported by morphometric, molecular/phylogenetic analyses, and SEM analysis (this only for H. arii). Previous SEM investigations are not available; nevertheless, observed qualitative characters can be compared regardless of the particular technique used as they are not linked to any measurement unit. Our SEM investigation of H. arii (), which is the type species of the genus, showed the generic diagnostic characters (digitate haptor, armed with marginal hooks; two pairs of anchors and two bars; outer root of anchors usually not expanded into wings), and also species-specific characters such as the appendix on dorsal bar (Yamaguti Citation1953; Bychowsky & Nagibina Citation1969; Lim Citation1996).
Hamatopeduncularia arii, reported from A. jella in the present study, resembled previous descriptions in all aspects (Yamaguti Citation1953; Bychowsky & Nagibina Citation1969; Paperna Citation1977; Lim Citation1996) except for a few morphological and morphometric variations. Both ring pads and the appendix on the dorsal bar were observed in the present specimens. However, the ring pads were not mentioned by Yamaguti (Citation1953), Bychowsky and Nagibina (Citation1969) or Paperna (Citation1977), and the appendix on the dorsal bar was not observed by Lim (Citation1996). Hamatopeduncularia arii was first reported by Yamaguti in 1953 from Arius spp. from Borneo. In 1969, Bychowsky and Nagibina redescribed this species from Arius falcarius (now Arius arius), Arius leiotetocephalus (now Plicofollis nella) and Arius maculatus from the South China Sea. Paperna (Citation1977) found it on Arius thalassinus (now Netuma thalassina) from Kenya. Lim (Citation1996) reported this species from A. maculatus, with characters similar to those of the original description except for the presence of ring pads on the inner root of the ventral anchors. As for the Indian region, Gupta and Khullar (Citation1967) reported H. arii from Arius sp.
Hamatopeduncularia elongatum was reported from A. jella in the present study and found to be in congruence with the description provided for the species by Lim (Citation1996) (i.e. nature of copulatory tube and haptor armature). It was first reported by Lim (Citation1996) from the gills of N. thalassina from the Strait of Malacca, off Peninsular Malaysia. In the present study, H. elongatum is for the first time reported from A. jella caught along Visakhapatnam coast, Bay of Bengal, India.
Hamatopeduncularia thalassini was reported from the host A. jella and first described by Bychowsky and Nagibina (Citation1969) from Tachysurus thalassinus (now N. thalassina) from the South China Sea (Hainan Island); later Paperna (Citation1977) briefly described this species with a few minor differences from the original account of Arius sp. Eventually, Lim (Citation1996) provided a complete description of the parasite, again from N. thalassina.
Hamatopeduncularia bifida sp. nov. resembles H. wallagonius Singh et al. 1995, H. malayanus Lim, Citation1996 and H. isosimplex Lim, Citation1996 in the general appearance of morphological characters, but differs from them in possessing a “V”-shaped dorsal bar and a straight ventral bar slightly bent in the centre, vs “V”-shaped dorsal and ventral bars. It further differs in the structure of the accessory piece, which is not reported as bifid in other forms. Besides, the length of the peduncle connecting the proper body with the haptor is considerably long in the present specimen. Interestingly, H. bifida sp. nov. formed a cluster with H. elongatum in the morphometric analysis, though a lot of variation is seen in the copulatory structure ( and ). Based on the differences noted with other species of the genus reported so far, the present species is considered a new species.
Hamatopeduncularia madhaviae sp. nov. from the host P. dussumieri differs from other species in the shape and structure of its copulatory organ. However, it shows some similarities with H. papernai Lim, Citation1995 and H. ritai Rastogi et al., Citation2005 in characters such as anchors, ventral bar, copulatory tube and marginal hooks, and their arrangement. The copulatory tube is slightly bent at the distal end in both H. papernai and H. madhaviae sp. nov. However, the only difference between the latter two species is the proximal end of the copulatory tube that is “C” (hook) shaped in H. madhaviae sp. nov., and closed cone-shaped in H. papernai. It differs from H. ritai in having an almost straight dorsal bar slightly bent in the middle, and in the absence of accessory piece and sleeve sclerites, whereas in H. ritai two accessory pieces and dorsal anchors with sleeve sclerites are present, and the dorsal bar is “V” shaped. As the present species showed significant morphological variation from other species reported so far, it is considered a new species.
Morphometric analysis
Based on morphometric data, both Euclidian analysis () and PCA () differentiated the five investigated Hamatopeduncularia species, H. arii, H. thalassini, H. bifida sp. nov. and H. elongatum (all found on the host A. jella), and H. madhaviae sp. nov. (retrieved on P. dussumieri), into three different clusters. Thus, the present morphometric study provides a solid basis for the separation of species within the same genus.
The structure of both haptoral and copulatory apparatus helped in defining these three groups. Within each of the two groups comprising two parasite species, all members exhibited similarities in their haptoral apparatus structures and clustered together in both the Euclidean cluster analysis and PCA: H. arii and H. thalassini (Group II) have similar haptoral sclerite morphologies; H. bifida sp. nov. and H. elongatum (Group I) show similar length of hooks and anchors. Nevertheless, within each of these two groups the two species of parasites can be easily distinguished by differences in the structure of the copulatory apparatus. Moreover, H. madhaviae sp. nov. from the host P. dussumieri, belonging to the third, separate cluster (Group III), showed differences as well in both its haptoral sclerite dimensions and copulatory apparatus structure.
Previous studies indicated that the haptoral morphologies of species are generally associated with attachment site on the host (niche) (Simkova et al. Citation2002). In the present study, two out of the four species found on A. jella (i.e. H. arii and H. thalassini) with similar haptoral morphology clustered together in the phylogenetic analysis, whereas the other two (H. elongatum and H. bifida sp. nov.), with similar haptoral sclerites, formed a second, separate cluster. Thus, it is suggested that this difference might be related to their specific niche on the host gill, i.e. attachment specificity on a particular area of the host gill.
Our cluster analysis identified an interesting set of associations when grouping parasite species according to their haptoral sclerite variability. Attachment organs of parasites, especially ectoparasites, have been considered important for host specificity. The monogenoidean haptor has been found to be highly specialised and therefore could constrain the parasites to certain sites on their hosts’ gills (Rohde Citation1989; Lambert & El Gharbi Citation1995). The present study supports the hypothesis that phylogenetically close parasite species may possess similar attachment organs, which might serve to better adapt them to their hosts. The morphology of two sclerotised organs, namely the attachment organ (the haptor) and the reproductive organ (including the copulatory piece and the vagina), mainly helps in the identification of monogenoidean parasites. The morphology of the haptor is considered useful for parasite determination at the generic level, while the reproductive organ seems to be more suitable for identification at the specific level, probably because of its higher rate of change (Pouyaud et al. Citation2006; Wu et al. Citation2007, Citation2008).
Molecular analysis
In the present study, morphological identification of the investigated Hamatopeduncularia species was integrated with their molecular characterisation, through the analysis of a molecular marker; this also allowed the study of the evolutionary relationships among these parasites. The nucleotide sequences of the parasites herein provided are the first 18S rDNA sequences ever reported for this genus. The 18S rDNA marker represents an excellent tool for both diagnostic purposes and phylogenetic studies (Lockyer et al. Citation2003; Buchmann & Bresciani Citation2006; Simkova et al. Citation2007; Wu et al. Citation2007; Mendlova et al. Citation2010; Shinn et al. Citation2010; Gilmore et al. Citation2012; Prikrylova et al. Citation2013; Müller et al. Citation2016). In our case, the 18S rDNA sequence analysis helped in both species discrimination and phylogenetic positioning, supporting and reflecting the classification based solely on morphological characters.
The new species H. elongatum and H. bifida sp. nov. significantly differ from each other in their copulatory organ structures; nevertheless, they show almost 100% identity in their 18S rDNA sequences. This discordance between morphological and molecular data raised some doubts in assigning the status of novel species to H. bifida. Three hypotheses can be formulated as possible explanations for this discrepancy: (1) the 18S rRNA gene is not the proper molecular marker for species identification between H. elongatum and H. bifida, i.e. the resolution power of this marker gene is low (this would support the existence of two different species); (2) the copulatory organ does not play an important role in taxonomical identification of the two species in analysis, so they should be considered the same species, i.e. H. elongatum; (3) an artefact occurred in laboratory analysis, i.e. specimens of the same species were erroneously collected and processed for molecular studies.
The second hypothesis, of course, is not in agreement with the traditional systematics considering hard-sclerotised structures, such as copulatory organs and haptoral sclerites, the taxonomically most important feature in distinguishing species in this group of parasites (Paperna Citation1979; Whittington Citation2005; Bruno et al. Citation2006; Whittington & Chisholm Citation2008; Pugachev et al. Citation2009).
If the third hypothesis were true, the two species would be obviously valid but the two 18S rRNA gene sequences could not be unambiguously attributed to any of them. Unfortunately, we did not have the opportunity to repeat the sampling (and the analysis), because the project was already finished at the time the molecular analysis was performed. On the other hand, we have no real hint that such an error occurred. Therefore, we decided to highlight this point in the hope that further studies will succeed in clarifying this question.
In the present paper we consider the first hypothesis more reliable. Consequently, we propose H. bifida as a true novel species, but we encourage further study using a combination of morphological and molecular techniques to specifically address the reliability of sclerotised structures as species-specific key features.
Consideration on the phylogeny of Hamatopeduncularia species
The taxonomic status of Hamatopeduncularia Yamaguti, Citation1953 has been discussed thoroughly by various authors (Kritsky & Boeger Citation1989; Lim Citation1996; Lim et al. Citation2001). In the original classification (Bychowsky Citation1937), the genus was placed in the order Dactylogyridea, family Dactylogyridae, subfamily Ancyrocephalinae, based on the structure of the copulatory organ and the armature of the haptor.
Gusev (Citation1961) accommodated all the known dactylogyridean genera from the siluriforms within the new subfamily Ancylodiscoidinae. Subsequently, Gusev (Citation1976) reassigned Hamatopeduncularia to the subfamily Ancyrocephalinae based on the type of seminal vesicle. Bychowsky and Nagibina (Citation1978) raised the Ancyrocephalinae to family status and re-assigned all dactylogyrids with four anchors (Ancyrocephalinae and Ancylodiscoidinae) to this group (Ancyrocephalidae), but later Kritsky and Boeger (Citation1989) rejected Ancyrocephalidae as a non-monophyletic taxon.
According to the World Register of Marine Species (WoRMS 2018, Hamatopeduncularia Yamaguti, Citation1953, http://www.marinespecies.org/aphia.php?p=taxdetails&id=517937 accessed 4 November 2018), the genus Hamatopeduncularia belongs to the family Ancylodiscoididae. According to our molecular analysis, Hamatopeduncularia is monophyletic and groups together with genera of the family Dactylogyridae, being the sister clade of Bychowskyella and Mizelleus. Monogenoids of all three genera parasitise catfishes belonging to the same order, i.e. Siluriformes, and show the dactylogyrid type of seminal vesicle (Gusev Citation1976).
Thus, considering both morphological and molecular results, we confirm that Hamatopeduncularia belongs to the family Dactylogyridae. Moreover, our phylogeny results are coherent with those provided by Mendoza-Franco and colleagues (Mendoza-Franco et al. Citation2018), whose analysis placed the 28S rDNA sequence of Hamatopeduncularia bagrae in association with the genera Thaparocleidus, Quadricanthus and Bichowskyella, within Dactylogyridae.
The monophyly of the family Dactylogyridae is well supported by high statistical value (1.00/100) in the present study. In our analysis, Ancyrocephalinae proved to be a polyphyletic taxon, with members scattered among different clades of Dactylogyridae (Klassen Citation1994; Lim Citation1998; Mollaret et al. Citation2000; Simkova et al. Citation2003, Citation2006; Plaisance et al. Citation2004, Citation2005; Mendoza-Palmero et al. Citation2015; Müller et al. Citation2016), and Pseudodactylogyrinae appeared to be non-monophyletic. Future studies are needed to address the issue of non-monophyly of subfamilies highlighted by our phylogenetic analysis based on the molecular marker 18S rDNA; an integrative morphological–molecular study approach is recommended.
Acknowledgements
The authors would like to thank Simone Gabrielli for figure elaboration.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Acosta AA, Franceschini L, Zago AC, Scholz T, Da Silva RJ. 2017. Six new species of Heteropriapulus (Monogenea: Dactylogyridae) from South American fishes with an amended diagnosis to the genus. Zootaxa 4290:459–482. DOI: 10.11646/zootaxa.4290.3.3.
- Acosta AA, Scholz T, Blasco-Costa I, Vieira Alves P, Da Silva RJ. 2018. A new genus and two new species of dactylogyrid monogeneans from gills of Neotropical catfishes (Siluriformes: Doradidae and Loricariidae). Parasitology International 67:4–12. DOI: 10.1016/j.parint.2017.09.012.
- Andreoli I, Mangini L, Ferrantini F, Santangelo G, Verni F, Petroni G. 2009. Molecular phylogeny of unculturable Karyorelictea (Alveolata, Ciliophora). Zoologica Scripta 38:651–662. DOI: 10.1111/j.1463-6409.2009.00395.x.
- Blair D, Barker SC. 1993. Affinities of the Gyliauchenidae: Utility of the 18S rRNA gene for phylogenetic inference in the digenea (Platyhelminthes). International Journal for Parasitology 23:527–532. DOI: 10.1016/0020-7519(93)90042-W.
- Bruno DW, Nowak B, Elliott DG. 2006. Guide to the identification of fish protozoan and metazoan parasites in stained tissue sections. Diseases of Aquatic Organisms 70:1–36. DOI: 10.3354/dao070001.
- Buchmann K, Bresciani J. 2006. Monogenea (Phylum Platyhelminthes). In: Woo PTK, editor. Fish diseases and disorders, volume 1: protozoa and metazoan infections 2th. Canada: University of Guelph, CABI. pp. 297–344. DOI:10.1079/9780851990156.0297
- Bychowsky BE. 1937. Ontogenesis and phylogenetic interrelationships of parasitic flat worms, Izvestiya Akademiya Nauk SSSR, Ser. Biologiya 4:1353–1383. (In Russian. English translation, 1967, Gloucester Point, Virginia Institute of Marine Sciences, Translation Series N°. 17)
- Bychowsky BE, Nagibina LF. 1969. Ancyrocephalinae (Dactylogyridae, Monogenoidea) from fishes of the family Ariidae. Parazitologiya 3:357–368. (In Russian with English summary)
- Bychowsky BE, Nagibina LF. 1978. A revision of Ancyrocephalinae Bychowsky, 1937 (Monogenoidea). Parazitologicheskii Sbornik 28:5–15. (In Russian)
- Caffara M, Locke SA, Cristanini C, Davidovich N, Markovich MP, Fioravanti ML. 2016. A combined morphometric and molecular approach to identifying metacercariae of Euclinostomum heterostomum (Digenea: Clinostomidae). The Journal of Parasitology 102:239–248. DOI: 10.1645/15-823.
- Castelli M, Sabaneyeva E, Lanzoni O, Lebedeva N, Floriano AM, Gaiarsa S, Benken K, Modeo L, Bandi C, Potekhin A, Sassera D, Petroni G 2018a. The extracellular association of the bacterium “Candidatus Deianiraea vastatrix” with the ciliate Paramecium suggests an alternative scenario for the evolution of Rickettsiales. BioRxiv 479196. DOI: 10.1101/479196.
- Castelli M, Serra V, Senra MVX, Basuri CK, Soares CAG, Fokin SI, Modeo L, Petroni G. 2018b. The hidden world of Rickettsiales symbionts: “Candidatus Spectririckettsia obscura”, a novel bacterium found in Brazilian and Indian Paramecium caudatum. Microbial Ecology. DOI:10.1007/s00248-018-1243-8.
- Chiellini C, Pasqualetti C, Lanzoni O, Fagorzi C, Bazzocchi C, Fani R, Petroni G, Modeo L. 2019. Harmful effect of Rheinheimera sp. EpRS3 (Gammaproteobacteria) against the protist Euplotes aediculatus (Ciliophora, Spirotrichea): Insights into the ecological role of antimicrobial compounds from environmental bacterial strains. Frontiers in Microbiology 10:510. DOI: 10.3389/fmicb.2019.00510.
- Clarke KR, Green RH. 1988. Statistical design and analysis for a “biological effects” study. Marine Ecology Progress Series 46:213–226. DOI: 10.3354/meps046213.
- Cribb TH, Bray RA. 2010. Gut wash, body soak, blender and heat-fixation: Approaches to the effective collection, fixation and preservation of trematodes of fishes. Systematic Parasitology 76:1–7. DOI: 10.1007/s11230-010-9229-z.
- Darriba D, Taboada GL, Doallo R, Posada D. 2012. jModel Test 2: More models, new heuristics and parallel computing. Nature Methods 30 9:772. DOI:10.1038/nmeth.2109
- Domingues V, Geusivam BS, Watanabe A. 2016. Monogenoidea (Polyonchoinea: Dactylogyridae) parasitizing the gills of marine catfish (Siluriformes: Ariidae) inhabiting the Atlantic Amazon Coast of Brazil. Zootaxa 4127:301–326. DOI: 10.11646/zootaxa.4127.2.4.
- Ferrantini F, Fokin SI, Modeo L, Andreoli I, Dini F, Görtz H-D, Verni F, Petroni G. 2009. “Candidatus Cryptoprodotis polytropus,” a novel Rickettsia‐like organism in the ciliated protist Pseudomicrothorax dubius (Ciliophora, Nassophorea). Journal of Eukaryotic Microbiology 56: 119–129. DOI: 10.1111/jeu.2009.56.issue-2.
- Fisher ER, Hansen BT, Nair V, Hoyt FH, Dorward DW. 2012. Scanning electron microscopy. Current Protocols in Microbiology 25:2B.2.1-2B.2.47. DOI: 10.1002/9780471729259.mc02b02s25.
- Fokin SI, Serra V, Ferrantini F, Modeo L, Petroni G. 2019. “Candidatus Hafkinia simulans” gen. nov., sp. nov., a novel Holospora-like bacterium from the macronucleus of the rare brackish water ciliate Frontonia salmastra (Oligohymenophorea, Ciliophora): Multidisciplinary characterization of the new endosymbiont and its host. Microbial Ecology 1–15. DOI: 10.1007/s00248-018-1311-0.
- Foottit RG, Sorensen JT. 1992. Ordination in the study of morphology, evolution and systematics of Insects: Applications and quantitative genetic rationales. Amsterdam: Elsevier. pp. 10.
- Francová K, Seifertová M, Blažek R, Gelnar M, Mahmoud ZN, Řehulková E. 2017. Quadriacanthus species (Monogenea: Dactylogyridae) from catfishes (Teleostei: Siluriformes) in eastern Africa: New species, new records and first insights into interspecific genetic relationships. Parasites and Vectors 10:361. DOI: 10.1186/s13071-017-2223-4.
- Gerasev PI. 1992. Types of accessory bars in Dactylogyrus, functional and systematic significance. Journal of Parasitology 25:219–227.
- Gilmore SR, Cone DK, Lowe G, King SK, Jones SRM, Abbott CL. 2012. Molecular phylogeny of Gyrodactylus (Monogenea) parasitizing fishes in fresh water, estuarine, and marine habitats in Canada. Canadian Journal of Zoology 90:776–786. DOI: 10.1139/z2012-040.
- Guindon S, Gascuel O. 2003. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Systematic Biology 52:696–704. DOI: 10.1080/10635150390235520.
- Gupta NK, Khullar M. 1967. On three monogenetic trematodes from some Indian marine food fishes. Indian Journal of Helminthology 18:4.
- Gusev AV. 1961. New subfamily of monogeneans (Monogenoidea). Doklady Akademii Nauk SSSR 139:1480–1482. (in Russian).
- Gusev AV. 1976. Freshwater Indian Monogenoidea. Principles of systematics, analysis of world faunas and their evolution. Indian Journal of Helminthology 25/26:1–241.
- Gusev AV. 1977. Principles of construction of the system Monogenoidea by B.E. Bychowsky. Parazitologicheskii Sbornik 27:18–26. (in Russian)
- Kearn GC, Whittington LD. 1994. Ancyrocephaline monogeneans of the genera Chauhanellus and Hamatopeduncularia from the gills of the blue catfish, Arius graeffei, in the Brisbane river and Moreton Bay, Queensland, Australia with descriptions of four new species. International Journal for Parasitology 24:569–588. DOI: 10.1016/0020-7519(94)90149-X.
- Klassen GJ. 1994. Phylogeny of Haliotrema species (Monogenea, Ancyrocephalidae) from boxfishes (Tetraodontiformes, Ostraciidae) – Are Haliotrema species from boxfishes monophyletic? Journal of Parasitology 80:596–610. DOI: 10.2307/3283198.
- Kritsky DC, Boeger WA. 1989. The phylogenetic status of the Ancyrocephalidae Bychowsky, 1937 (Monogenea, Dactylogyroidea). Journal of Parasitology 75:207–211. DOI: 10.2307/3282767.
- Lambert A, El Gharbi S. 1995. Monogenean host specificity as a biological and taxonomic indicator for fish. Biological Conservation 72:227–235. DOI: 10.1016/0006-3207(94)00085-5.
- Lanzoni O, Sabaneyeva E, Modeo L, Castelli M, Lebedeva N, Verni F, Schrallhammer M, Potekhin A, Petroni G. 2019. Diversity and environmental distribution of the cosmopolitan endosymbiont “Candidatus Megaira”. Scientific Reports 9:1179. DOI: 10.1038/s41598-018-37629-w.
- Lim LHS. 1995. Neocalceostoma Tripathi, 1957 and Neocalceostoma Krisky, Mizelle & Bilquees, 1978: (Monogenea: Neocalcestoma n. fam.) from ariid fishes of Peninsular Malaysia. Systematic Parasitology 30:141–151. DOI: 10.1007/BF00010167.
- Lim LHS. 1996. Eight species of Hamatopeduncularia Yamaguti, 1953 (Monogenea: Ancyrocephalidae) from Ariidae of Peninsular Malaysia. Systematic Parasitology 33:53–71. DOI: 10.1007/BF00009720.
- Lim LHS. 1998. Diversity of monogeneans in Southeast Asia. International Journal for Parasitology 28:1495–1515. DOI: 10.1016/S0020-7519(98)00061-7.
- Lim LHS, Timofeeva TA, Gibson DI. 2001. Dactylogyridean monogeneans of the siluriform fishes of the old world. Systematic Parasitology 50:159–197. DOI: 10.1023/A:1012237801974.
- Lockyer AE, Olson PD, Littlewood DTJ. 2003. Utility of complete large and small subunit rRNA genes in resolving the phylogeny of the Neodermata (Platyhelminthes): Implications and a review of the cercomer theory. Biological Journal of the Linnean Society 78:156–171. DOI: 10.1046/j.1095-8312.2003.00141.x.
- Ludwig W, Strunk O, Westram R, Richter L, Meier H, Yadhukumar H. 2004. ARB: A software environment for sequence data. Nucleic Acids Research 32:1363–1371. DOI: 10.1093/nar/gkh293.
- Medlin L, Elwood HJ, Stickel S, Sogin ML. 1988. The characterization of enzymatically amplified 16S-like rRNA- coding regions. Gene 71:491–499. DOI: 10.1016/0378-1119(88)90066-2.
- Mendlova M, Pariselle A, Vyshocilova M, Simkova A. 2010. Molecular phylogeny of monogeneans parasitizing African freshwater Cichlidae inferred from LSU rDNA sequences. Parasitology Research 107:1405–1413. DOI: 10.1007/s00436-010-2008-6.
- Mendoza-Franco EF, Rosado TMC, Duarte AAD, Rodríguez RER. 2018. Morphological and molecular (28S rRNA) data of monogeneans (Platyhelminthes) infecting the gill lamellae of marine fishes in the Campeche Bank, southwest Gulf of Mexico. ZooKeys 783:125–161. DOI: 10.3897/zookeys.783.26218.
- Mendoza-Palmero CA, Blasco-Costa I, Scholz T. 2015. Molecular phylogeny of Neotropical monogeneans (Platyhelminthes: Monogenea) from catfishes (Siluriformes). Parasites & Vectors 8:164. DOI: 10.1186/s13071-015-0767-8.
- Modeo L, Fokin SI, Boscaro V, Andreoli I, Ferrantini F, Rosati G, Verni F, Petroni G. 2013a. Morphology, ultrastructure, and molecular phylogeny of the ciliate Sonderia vorax with insights into the systematics of order Plagiopylida. BMC Microbiology 13:40. DOI: 10.1186/1471-2180-13-40.
- Modeo L, Petroni G, Lobban CS, Verni F, Vannini C. 2013b. Morphological, ultrastructural, and molecular characterization of Euplotidium rosati n. sp. (Ciliophora, Euplotida) from Guam. Journal of Eukaryotic Microbiology 60:25–36. DOI: 10.1111/jeu.12003.
- Mollaret I, Jamieson BGM, Justine JL. 2000. Phylogeny of the Monopisthocotylea and Polyopisthocotylea (Platyhelminthes) inferred from 28S rDNA sequences. International Journal for Parasitology 30:171–185. DOI: 10.1016/S0020-7519(99)00197-6.
- Müller MI, Ceccarelli PS, Ueta MT. 2016. Supplementary studies on Anacanthorus penilabiatus and Mymarothecium viatorum (Monogenea: Dactylogyridae) from Piaractus mesopotamicus (Characiformes: Serrasalmidae) in Brazil. Acta Parasitologica 61:508–515. DOI: 10.1515/ap-2016-0067.
- Nitla V, Serra V, Fokin SI, Modeo L, Verni F, Sandeep BV, Kalavati C, Petroni G. 2018. Critical revision of the family Plagiopylidae (Ciliophora: Plagiopylea), including the description of two novel species, Plagiopyla ramani and Plagiopyla narasimhamurtii, and redescription of Plagiopyla nasuta Stein, 1860 from India. Zoological Journal of the Linnean Society. DOI: 10.1093/zoolinnean/zly041/5095288.
- Oliver G 1987. Les Diplectanidae Bychowsky, 1957 (Monogenea, Monopisthocotylea, Dactylogyridae). Systématique. Biologie. Ontogénie. Écologie. Essai de phylogenèse. Thèse d’État, Académie de Montpellier, Université des Sciences et Technique du Languedoc, Montpellier. 434 pp.
- Paperna I. 1977. The Monogenea of marine catfish. Sobretiro Excerta De Parasitologica in Memoria Del Doctor Eduardo Caballeroy, Instituto De Biologia, Universidad Nacional Autonoma De Mexico, Publicaciones Especiacles 4:99–116.
- Paperna I. 1979. Monogenea of Inland Water Fish in Africa. Annalen Koninklijk Museum voor Midden-Afrika, Zoologische Wetenschappen Tervuren 226:1–131.
- Petroni G, Dini F, Verni F, Rosati G. 2002. A molecular approach to the tangled intrageneric relationships underlying Phylogeny in Euplotes (Ciliophora, Spirotrichea). Molecular Phylogenetics and Evolution 22:118–130. DOI: 10.1006/mpev.2001.1030.
- Plaisance L, Bouamer S, Morand S. 2004. Description and redescription of Haliotrema species (Monogenoidea: Polyonchoinea: Dactylogyridae) parasitizing butterfly fishes (Teleostei: Chaetodontidae) in the Indo-West Pacific Ocean. Parasitology Research 93:72–78. DOI: 10.1007/S00436-004-1094-8.
- Plaisance L, Littlewood DTJ, Olson PD, Morand S. 2005. Molecular phylogeny of gill monogeneans (Platyhelminthes, Monogenea, Dactylogyridae) and the colonization of Indo- West Pacific butterflyfish hosts (Perciformes, Chaetodontidae). Zoologica Scripta 34:425–436. DOI: 10.1111/j.1463-6409.2005.00191.x.
- Pouyaud L, Desmarais E, Deveney M, Pariselle A. 2006. Phylogenetic relationships among monogenean gill parasites (Dactylogyridea, Ancyrocephalidae) infesting tilapiine hosts (Cichlidae): Systematic and evolutionary implications. Molecular Phylogenetics and Evolution 38:241–249. DOI: 10.1016/j.ympev.2005.08.013.
- Prikrylova I, Vanhove MPM, Janssens SB, Billeter PA, Huyse T. 2013. Tiny worms from a mighty continent: High diversity and new phylogenetic lineages of african monogeneans. Molecular Phylogenetics and Evolution 67:43–52. DOI: 10.1016/j.ympev.2012.12.017.
- Pruesse E, Quast C, Knittel K, Fuchs BM, Ludwig W, Peplies J, Glockner FO. 2007. SILVA: A comprehensive online resource for quality checked and aligned ribosomal RNA sequence data compatible with ARB. Nucleic Acids Research 35:7188–7196. DOI: 10.1093%2Fnar%2Fgkm864.
- Pugachev ON, Gerasev PI, Gussev AV, Ergens R, Khotenowsky I. 2009. Guide to Monogenoidea of freshwater fish of Palaearctic and Amur regions. Milan: Ledizione-The Innovative Ledi Publishing Company. pp. 564.
- Rastogi P, Arya PV, Singh HS. 2005. On three new species of genus Hamatopeduncularia Yamaguti, 1963, from freshwater fishes of Meerut, with a note on its biogeography. Journal of Experimental. Zoology India 8:205–220.
- Rohde K. 1989. Simple ecological systems, simple solutions to complex problems? Evolutionary Theory 8:305–350.
- Ronquist F, Teslenko M, Van der Mark P, Ayres DL, Darling A, Höhna S, Larget B, Liu L, Suchard MA, Huelsenbeck JP. 2012. Mrbayes 3.2: Efficient bayesian phylogenetic inference and model choice across a large model space. Systematic Biology 61:539–542. DOI: 10.1093/sysbio/sys029.
- Rosati G, Modeo L, Melai M, Petroni G, Verni F. 2004. A multidisciplinary approach to describe protists: A morphological, ultrastructural, and molecular study on Peritromus kahli Villeneuve-Brachon, 1940 (Ciliophora, Heterotrichea). Journal of Eukaryotic Microbiology 51: 49–59. DOI:10.1111/j.1550-7408.2004.tb00160.x.
- Sailaja B, Shameem U, Madhavi R. 2016. Redescription and morphometric analysis of Paramazocraes thrissocles Tripathi, 1959 and P-setipinna Zhang & Ding in Zhang, Yang & Liu, 2001 (Monogenea: Mazocraeidae) infecting clupeoid fishes off Visakhapatnam coast, Bay of Bengal. Systematic Parasitology 93:193–203. DOI: 10.1007/s11230-015-9616-6.
- Shinn AP, Collins C, Garcia-Vasquez A, Snow M, Matejusova I, Paladini G, Longshaw M, Lidenstrom T, Stone DM, Turnbull JF, Picon-Camacho PM, Vasquez-Rivera C, Duguid RA, Mo TA, Hansen H, Olstad K, Cable J, Harris PD, Kerr R, Graham D, Monagham SJ, Yoon GH, Buchmann K, Taylor NGH, Bakke TA, Raynard R, Irving S, Bron JE. 2010. Multi-centre testing and validation of current protocols for the identification of Gyrodactylus salaris (Monogenea). International Journal for Parasitology 40:1455–1467. DOI: 10.1016/j.ijpara.2010.04.016.
- Simkova A, Matejusova I, Cunningham CO. 2006. A molecular phylogeny of the Dactylogyridae sensu Kritsky and Boeger (1989) (Monogenea) based on the D1-D3 domains of large subunit rDNA. Parasitology 133:43–53. DOI: 10.1017/S0031182006009942.
- Simkova A, Ondračkova M, Gelnar M, Morand S. 2002. Morphology and coexistence of congeneric ectoparasite species: Reinforcement of reproductive isolation? Biological Journal of the Linnean Society of London 76:125–135. DOI: 10.1111/j.1095-8312.2002.tb01719.x.
- Simkova A, Pecinkova M, Rehulkova E, Vyskocilova M, Ondrackova M. 2007. Dactylogyrus species parasitizing european Barbus species: Morphometric and molecular variability. Parasitology 134:1751–1765. DOI: 10.1017/S0031182007003265.
- Simkova A, Plaisance L, Matejusova I, Morand S, Verneau O. 2003. Phylogenetic relationships of the Dactylogyridae Bychowsky, 1933 (Monogenea: Dactylogyridae): The need for the systematic revision of the Ancyrocephalinae Bychowsky, 1937. Systematic Parasitology 54:1–11. DOI: 10.1023/A:1022133608662.
- Strona G, Montano S, Seveso D, Galli P, Fattorini S. 2013. Identification of Monogenea made easier: A new statistical procedure for an automatic selection of diagnostic linear measurements in closely related species. Journal of Zoological Systematics and Evolutionary Research 52:95–99. DOI: 10.1111/jzs.12050.
- Sudarsan D, John ME, Somavanshi VS. 1990. Marine fishery resources potential in the Indian exclusive economic zone — An update. Mumbai: Fishery Survey of India.
- Takemoto RM, Amato JFR, Luque JL. 1996. Comparative analysis of the metazoan parasite communities of leatherjackets, Oligoplites palometa, O. saurus and O. saliens (Osteichthyes: Carangidae) from Sepetiba Bay, Rio de Janeiro, Brazil. Revista Brasileira de Biologia 56:639–650.
- Tedesco P, Gustinelli A, Caffara M, Patarnello P, Terlizzi A, Fioravanti ML. 2018. Hysterothylacium fabri (Nematoda: Raphidascarididae) in Mullus surmuletus (Perciformes: Mullidae) and Uranoscopus scaber (Perciformes: Uranoscopidae) from the Mediterranean. Journal of Parasitology 104:262–274. DOI:10.1645/17-115
- Tripathi YR. 1959. Monogenetic trematodes from fishes of India. Indian Journal of Helminthology 9:1–149.
- Verma C, Chaudhary A, Singh HS. 2017b. Redescription and phylogenetic analyses of Thaparocleidus gomtius and T. sudhakari (Monogenea: Dactylogyridae) from Wallago attu (Siluriformes: Siluridae) in India. Helminthologia 54:87–96. DOI: 10.1515/helm-2017-0008.
- Verma C, Chaudhary A, Singh HS. 2017c. Morphology, molecular and systematic analyses of Bychowskyella (Monogenea: Dactylogyridae) in siluriform fish from India. Journal of Helminthology 91:197–205. DOI: 10.1017/S0022149X16000122.
- Verma J, Agrawal N, Verma AK. 2017a. The use of large and small subunits of ribosomal DNA in evaluating phylogenetic relationships between species of Cornudiscoides Kulkarni, 1969 (Monogenoidea: Dactylogyridae) from India. Journal of Helminthology 91:206–214. DOI: 10.1017/S0022149X16000134.
- Whittington ID. 2005. Monogenea Monopisthocotylea (ectoparasitic flukes). In: Rohde K, editor. Marine Parasitology. Wallingford: CABI Publishing. pp. 63–72.
- Whittington ID, Chisholm LA. 2008. Diseases caused by Monogenea. In: Eiras J, Segner H, Wahli T, Kapoor BG, editors. Fish diseases. Enfield: Science Publishers. pp. 683–816.
- Wu XY, Zhu XQ, Xei MQ, Li AX. 2007. The evaluation for generic level monophyly of Ancyrocephalinae (Monogenea, Dactylogyridae) using ribosomal DNA sequence data. Molecular Phylogenetics and Evolution 44:530–544. DOI: 10.1016/j.ympev.2007.03.025.
- Wu XY, Zhu XQ, Xie MQ, Wang JQ, Li AX. 2008. The radiation of Thaparocleidus (Monogenoidea: Dactylogyridae: Ancylodiscoidinae): Phylogenetic analyses and taxonomic implications inferred from ribosomal DNA sequences. Parasitology Research 102:283–288. DOI: 10.1007/s00436-007-0760-z.
- Yamada PDOF, Yamada FH, Da Silva RJ, Dos Anjos LA. 2017. A new species of Cosmetocleithrum (Monogenea, Dactylogyridae), a gill parasite of Trachelyopterus galeatus (Siluriformes, Auchenipteridae) from Brazil, with notes on the morphology of Cosmetocleithrum striatuli. Comparative Parasitology 84:119–123. DOI: 10.1654/1525-2647-84.2.119.
- Yamaguti S. 1953. Parasitic worms mainly from the Celebes. Part 2. Monogenetic trematodes of fishes. Acta Medicinae Okayama 5:257–295.
- Yao W, Wang W, Xia X, Chen W. 1998. Four new species of Ancyrocephalidae of marine fishes from Nanao Island, Guangdong. Acta Hydrobiologica Sinica 22:120–125. In Chinese
- Zhu X, Gasser RB, Chilton NB. 1998. Differences in the 5.8S rDNA sequences among ascarid nematodes. International Journal for Parasitology 28:617–622. DOI: 10.1016/S0020-7519(97)00214-2.