Abstract
The actual occurrence of Squalus megalops in the Mediterranean Sea has recently been questioned. Several research works which sought to assess available morphological and meristic features that differentiate S. megalops from other Squalus species in the Mediterranean Sea, revealed poor discriminatory power and high variability of the assessed characters, especially when comparing S. megalops and S. blainville. The application of molecular tools does not support the presence of S. megalops. In the present study, we screened spurdog species from the Strait of Sicily using a molecular taxonomy approach based on two mitochondrial DNA markers and we report the occurrence of two Squalus lineages characterizing specimens collected from the stretch of sea between Tunisia, southern Sicily, Malta and Libya. The results support the hypothesis that a common species, S. blainville, currently inhabits the Mediterranean Sea, while a second and rare species is probably an occasional visitor with high morphological similarity to the S. megalops and S. blainville but is genetically distinct from both. Within this perspective, the occurrence of S. megalops in the Mediterranean Sea is not confirmed and our study highlights the taxonomic uncertainties in relation to the occurrence and distribution of Squalus species in this region. We encourage the establishment of a coordinated international effort to implement a comprehensive and integrated taxonomic assessment on this genus which represents an irreplaceable component of the biodiversity of the area.
Introduction
The intrinsic low variation of morphological characters specific to elasmobranchs hinders their taxonomic identification at the species level and consequently undermines their conservation at different geographical scales (McEachran & Dunn Citation1998; Quattro et al. Citation2006; Aschliman et al. Citation2012). The lack of well-preserved holotypes for many shark species (e.g. Centrophorus spp.), misidentifications in databases and in the literature, and challenges in retrieving representative series of specimens for comparison are top-down impediments to the proper taxonomic identification and the potential revision of genera (Veríssimo et al. Citation2014).
In particular, the genus Squalus Linnaeus, 1758 is distributed worldwide (Ebert & Stehmann Citation2013) and includes about 26 different species of long-lived sharks (Viana et al. Citation2016) inhabiting the waters of the continental shelf and upper slope, between 300–700 m of depth (Whitehead et al. Citation1984; Cannizzaro et al. Citation1995; Serena et al. Citation2009), as well as some seamounts and the waters around oceanic islands (Compagno Citation1984; Veríssimo et al. Citation2017). The genus is highly represented in bycatch and several studies have focused on the mitigation of the fishery impact on this group (Mandelman & Farrington Citation2007; Stoner & Kaimmer Citation2008; Tallack & Mandelman Citation2009). In addition, most spurdog shark species are included in the IUCN Red List of Threatened Species and are currently classified in categories from Data Deficient to Endangered (i.e. the Greeneye spurdog S. chloroculus Last et al. Citation2007; IUCN Citation2019). For this reason, highlighting the need to increase our knowledge about taxonomy, biology and ecology of these species appears now essential. Difficulties due to the problems of distinguishing between morphologically similar species and to the lack of effective and user-friendly identification field guides have been commented upon by many authors (Garrick Citation1960; Compagno Citation1984; Munoz-Chapuli & Ramos Citation1989; Veríssimo et al. Citation2017). With the progressive expansion of molecular taxonomy (i.e. DNA barcoding) and its integration with morphological approaches, unravelling the taxonomy of confounding groups of sharks has become a priority for their conservation (Ebert et al. Citation2010; Veríssimo et al. Citation2014; Pfleger et al. Citation2018). In more recent times, the well-established DNA barcoding technique based on the mitochondrial DNA (mtDNA) cytochrome c oxidase subunit I (COI) has been coupled with analysis of faster-evolving mitochondrial genes or long ribosomal DNA to improve the resolution and support of inferred phylogenetic relationship, up to the description of the evolutionary history of species (Avise Citation2004; Naylor et al. Citation2012; Krehenwinkel et al. Citation2019).
Recently, Veríssimo et al. (Citation2017) reported uncertainties in the identification of Squalus specimens caught along the eastern Atlantic Ocean and in the Mediterranean Sea. In the latter area, Squalus acanthias Linnaeus, 1758 and Squalus blainville (Risso, 1827) are considered as resident species, while the presence of S. megalops (MacLeay, 1881) has been reported from Tunisia (Marouani et al. Citation2012). From the latter region, several specimens were analysed with a multidisciplinary approach and significant differences were reported for the two identified groups defined respectively as S. blainville and S. megalops (Marouani et al. Citation2012). Sequence data from both identified species were included later in the comprehensive assessment of the Squalus genus carried out in Veríssimo et al. (Citation2017). Using two mtDNA markers, three main groups and four main Clades corresponding to S. acanthias (Clade A), S. blainville/S. megalops (Clade B), S. megalops (Clade C) and Squalus mitsukurii Jordan & Snyder, 1903 (Clade D) were identified in the Eastern Atlantic and the Mediterranean Sea (Veríssimo et al. Citation2017). Individuals classified under Clade C, which is highly divergent from both S. megalops from Australian waters and Clade B, originates from tropical West African coasts, with the exception of the single specimen from Tunisia, previously identified as S. blainville by Marouani et al. (Citation2012). The sequence data associated with the individual from Tunisian waters described as S. megalops in Marouani et al. (Citation2012) fitted in Clade B. Given the above described results and since specimens classified as potential S. megalops are included in both Clade B and Clade C, these findings further support current inconsistencies in species identification within the genus Squalus and the need for an accurate redescription of Squalus species, especially in the Mediterranean Sea, to stabilize the systematic and facilitate specimens identification.
Extensive studies conducted in other areas of the Mediterranean Sea considering both morphological (chondrocranium and other body traits) and genetic (COI sequences) analyses, revealed the presence of only one spurdog species, S. blainville, in the Ionian, Lybian, Aegean Seas and in the Sardinian waters (Kousteni et al. Citation2016; Bellodi et al. Citation2018). These findings spotlighted the stretch of sea between Tunisia, southern Sicily, Malta and Libya, known as the Strait of Sicily, as a more interesting area for spurdog species. Differently from Marouani et al. (Citation2012), Bonello et al. (Citation2016) did not identify diagnostic features (e.g. dermal denticles) which could effectively discriminate between S. blainville and S. megalops. The presence of S. blainville in the Maltese waters was assessed through the use of the DNA barcoding approach (Bonello et al. Citation2016). In the same region, Vella et al. (Citation2017) collected and analysed individuals belonging to the nominal S. blainville and genetically clustering within Clade B (sensu Veríssimo et al. Citation2017), while three individuals were classified as Squalus sp. by the authors as clustering in the genetic Clade C (sensu Veríssimo et al. Citation2017).
In the present study, we screened all spurdogs caught in the Strait of Sicily between 2016 and 2017 to investigate the species composition in this stretch of sea. Comparing new and publicly available mtDNA gene sequences (COI and NADH dehydrogenase subunit 2; NADH2) we investigated the possible co-occurrence of two or more species of spurdog within the study area. Furthermore, since previous studies revealed that nominal species characterized by a wide geographical distribution share mitochondrial lineages, we evaluated the relationship between the genetic lineages retrieved in the Strait of Sicily and other Clades previously identified within the genus Squalus.
Materials and methods
Sample collection and DNA extraction
A total of 160 individuals of Squalus where caught off the southern coast of Sicily (General Fisheries Commission for the Mediterranean Geographical Subarea, GFCM-GSA 16) between July 2016 and May 2017 at depths between 83 m and 452 m, in the framework of the International Bottom Trawl Survey in the Mediterranean programme (MEDITS, Spedicato et al. Citation2019) and the Campionamento Biologico (CAMP-BIOL) monitoring programme of commercial catch (Milisenda et al. Citation2017). Twelve additional samples of Squalus blainville caught around the Maltese Islands (GFCM-GSA 15) in 2012 (COI analyses included in Bonello et al. Citation2016) and ten specimens collected off the Tuscany coasts (GFCM-GSA 9) in 2017 were included in this study. All specimens were preserved at −20°C until the collection of main biological data and tissues samples for genetic analyses. In particular, individual muscle tissue or fin clips were preserved in 96% ethanol for laboratory analyses (see Table S1 for details on the field species assignment and geographical origin of specimens).
Total genomic DNA (gDNA) was extracted from approximately 20 mg of tissue using the Wizard® SV Genomic DNA Purification System by Promega, according to the manufacturer’s instructions. The quality of the extracted gDNA was assessed on a 0.8% agarose gel electrophoresis.
mtDNA amplification and sequencing
A fragment of the mitochondrial gene COI (~650 bp) was obtained from each specimen by PCR using the FishF2 and FishR2 primers (Ward et al. Citation2005). PCR reactions were performed in 25 μl total volume containing 2.5 μl gDNA template, 10 μl nuclease-free water, 5 μl 1x PCR buffer, 1.5 μl of MgCl2 (50 mM), 2 μl dNTPs (10 mM), 1.5 μl of each primer (10 μM) and 0.25 U GoTaq G2 Flexi DNA polymerase (Promega). Amplifications were performed in a T-gradient thermocycler (Biometra) with an initial denaturation of 2 min at 95°C, followed by 35 cycles of 30 s at 94°C, 30 s at 54°C and 60 s at 72°C and a final extension step for 7 min at 72°C.
Similarly, a fragment of the NADH2 (~1100 bp) gene was amplified for all individuals using the Met-F and Trp-R primers (Vella et al. Citation2017). PCR conditions consisted of an initial denaturation of 2 min at 95°C, followed by 28 cycles of 45 s at 95°C, 45 s at 54°C and 60 s at 72°C and a final extension step for 7 min at 72°C. PCR outcomes were evaluated on a 2% agarose gel and preserved at 4°C until purification. All PCR products were purified using the ExoSAP-IT™ Express PCR Product Cleanup Reagent (ThermoFisher Scientific) following the manufacturer’s protocol. All amplicons were then sequenced with the same primers used for the amplification by the external provider Macrogen Europe (Amsterdam, the Netherlands).
Data analysis
The mtDNA COI and NADH2 sequences electropherograms were imported in MEGA v.X (10.1; Kumar et al. Citation2018), and carefully assessed and edited. All the sequences for each marker were aligned with the CLUSTAL W algorithm (Thompson et al. Citation1994) as implemented in MEGA and the correct amino acidic translation was verified to exclude nuclear mitochondrial pseudogenes (Song et al. Citation2008).
The software DnaSP v.6 (Rozas et al. Citation2017) has been used to compute the number of haplotypes, the number of polymorphic and parsimony-informative sites, the haplotype (Hd) and nucleotide diversity (Pi) were estimated for each of the newly obtained COI and NADH2 datasets.
Available COI and NADH2 sequences were retrieved for S. acanthias, Squalus albifrons Last et al. Citation2007, S. blainville, Squalus brevirostris Tanaka, 1917, Squalus chloroculus Last et al. Citation2007, Squalus clarkae Pfleger et al. Citation2018, Squalus crassispinus Last, Edmunds & Yearsley, 2007, Squalus cubensis Howell Rivero, 1936, Squalus edmundsi White, Last & Stevens, 2007, Squalus formosus White and Iglesias (Citation2011), Squalus grahami White, Last & Stevens, 2007, Squalus griffini Phyllipps, 1931, Squalus hemipinnis White, Last & Yearsley, 2007, Squalus japonicus Ishikawa, 1908, S. megalops, S. mitsukurii, Squalus montalbani Whiteley, 1931, Squalus nasutus Last, Marshall & White, 2007, Squalus raoulensis Duffy & Last, 2007 and Squalus suckleyi (Girard, 1855) from both BOLDsystems v.4 (Ratnasingham & Hebert Citation2007; http://www.boldsystems.org) and NCBI databases (https://www.ncbi.nlm.nih.gov) and added to the original dataset (see Table S2). In both datasets, when possible, retrieved data had different geographic origin to properly assess intraspecific variability.
For each mtDNA marker, a Neighbour-joining (NJ) tree (Saitou & Nei Citation1987) was computed with MEGA using p-distance (Collins & Cruickshank Citation2013) and, although no nucleotide gap was detected across both datasets, the ‘pairwise deletion’ option for the treatment of gaps and missing data was selected. A bootstrap test (BS) with 10,000 replicates (Felsenstein Citation1985) was performed to evaluate the robustness of reconstructions. The average intra and interspecific genetic distances among the clades observed were calculated with MEGA.
In order to obtain more robust and comparable results in relation to previous studies (Marouani et al. Citation2012; Bonello et al. Citation2016; Vella et al. Citation2017; Veríssimo et al. Citation2017), a concatenated dataset was created merging the newly obtained COI and NADH2 individual sequences with those published by Veríssimo et al. (Citation2017) and Vella et al. (Citation2017). The two mtDNA genes were concatenated in a congruent dataset (see Table S2), using Phyutility v.2.2 (Smith & Dunn Citation2008). A second NJ tree was computed on the concatenated dataset as reported above.
The software IQ-TREE v.1.6.12 (Nguyen et al. Citation2015) was firstly used to identify the best substitution model to be applied to the concatenated dataset and then to compute the Maximum-Likelihood (ML) reconstruction using the TN+F+ G4 model and an ultrafast BS (Hoang et al. Citation2018) with 10,000 replicates.
The Bayesian Inference (BI) reconstruction was unravelled with MrBayes v.3.2.7 (Ronquist et al. Citation2012) using two independent runs of 1,000,000 generations and a 25% burn-in cut-off. Run convergence was assessed considering a mean standard deviation of split frequencies of <0.01 between runs.
For all the topology reconstruction methods, Cirrhigaleus asper (Merrett, 1973) (COI and NADH2 Accession numbers MN982926 and JQ518974, respectively) and Cirrhigaleus australis White, Last & Stevens, 2007 (NC_024059) were chosen as outgroups for the analyses. The acquired trees were summarised in one congruent topology and edited using FigTree v.1.4.2 (Rambaut & Drummond Citation2012). BS values for NJ and ML trees were reported near nodes, as well as the posterior probability (P) obtained for the BI reconstruction.
The phylogeographic relationship of species haplotypes was inferred with the Median Joining Tree clustering algorithm (Bandelt et al. Citation1999) implemented in the software PopART (Leigh & Bryant Citation2015). The network was obtained considering all the Squalus spp. nominal species and lineages detected across the concatenated dataset, represented by S. acanthias, S. blainville, S. griffini, Squalus cf. megalops (Mauritius), Squalus cf. mitsukurii (Uruguay and Hawaii), Squalus sp. Clade B (sensu Veríssimo et al. Citation2017), Squalus sp. Clade C (sensu Veríssimo et al. Citation2017) and Squalus sp. Clade D (sensu Veríssimo et al. Citation2017).
Results
DNA was successfully extracted from all tissue samples, although PCR amplification and sequencing were successful for 121 and 107 individuals for COI and NADH2, respectively. These newly obtained sequences of Squalus specimens collected in the Strait of Sicily and Tuscany coasts, have been deposited in GenBank under the Accession Numbers (COI Accession Numbers MW537886-MW537998; NADH2 Accession Numbers MW557187-MW557293).
The new COI sequences counted a total of 51 polymorphic and 33 parsimony informative sites, 54 mutations and 33 haplotypes (Hd = 0.781 ± 0.034; Pi = 0.005 ± 0.027), while a total of 54 polymorphic and 33 parsimony informative sites, 56 mutations and 39 haplotypes (Hd = 0.874 ± 0.022; Pi = 0.006 ± 0.022) were detected among new NADH2 sequences.
After retrieving additional sequences from public data repositories, the final dataset for COI included 735 sequences (499 bp long) representing 18 nominal species, while the NADH2 dataset included 352 sequences (523 bp long) representing 20 nominal species. The concatenated dataset consisted of 222 sequences (1022 bp long).
Considering the congruence among the NJ, ML and BI tree topologies obtained using the concatenated dataset (Table S2), all reconstructions were summarised in one topology (), which assigned most of the individuals collected off the southern coast of Sicily, Malta and Tuscany to a bigger cluster including S. blainville from the Mediterranean Sea and Squalus sp. Clade B (sensu Veríssimo et al. Citation2017) from South Africa, Angola, Namibia, Morocco, Portugal and the Mediterranean Sea (BS = 100% for both NJ and ML and posterior probability, P = 1 for BI). Conversely, two specimens of Squalus (S7 and S9) are included within the bigger group composed by individuals of Squalus sp. collected in Gabon and Guinea, representing the genetic Clade C (sensu Veríssimo et al. Citation2017; BS = 100% for both NJ and ML and P = 1 for BI). Furthermore, the three Squalus sp. collected around Malta by Vella et al. (Citation2017) were grouped within Clade C ().
Figure 1. Tree topology based on concatenated mitochondrial sequences (COI and NADH2) summarising NJ, ML and BI reconstructions of the genus Squalus. BS values of NJ and ML and Bayesian posterior probability P are respectively reported top-down near nodes. BS values are reported when ≥70%. P is reported when ≥0.95. Non-supported nodes are indicated as a double dash. Clades including samples of Squalus collected in the Strait of Sicily are highlighted in grey
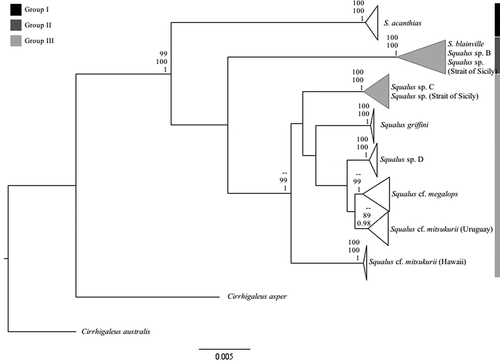
The NJ tree topologies reconstructed considering a large number of nominal species characterised by a wide geographic distribution (e.g. Australia, China, Indonesia, New Zealand, Japan, Taiwan, UK, USA) have consistently highlighted the existence of three distinct and well-supported groups (Figure S1, for COI; Figure S2, for NADH2). A first group (group I; Table S2) included the two main Clades of S. acanthias and S. suckleyi with high BS values in both COI and NADH2 reconstructions (100% and 99%, respectively), a second one (group II; Table S2) included the nominal S. blainville, S. brevirostris, S. megalops and S. raoulensis (BS = 100% in COI tree topology and BS = 99% in NADH2), and a last one (group III in Table S2) included all the other species considered with BS = 99% in COI.
Considering the COI mitochondrial gene, the genetic distance within groups was generally low, ranging from 0% (Squalus albifrons, S. grahami, S. griffini, Squalus cf. megalops, Squalus cf. mitsukurii from Hawaii) to 1.47% (S. nasutus) and 1.49% (S. megalops; Table S3). This was also confirmed by the NADH2 data, for which the genetic distance measured in S. mitsukurii complex sp. D and S. raoulensis was also 0%, while in S. brevirostris the genetic distance reached 3.02% (Table S4).
COI genetic distances between groups ranged from 0.10% (S. clarkae vs S. cubensis) to 7.90% (S. acanthias vs S. megalops or S. acanthias vs S. raoulensis; Table S3, for COI), while they ranged from 0% (S. mitsukurii complex sp. D vs Squalus sp. Clade D) and 8.78% (S. edmundsi vs S. suckleyi) for NADH2 (Table S4). COI genetic distances increased when comparing Squalus individuals caught in the Strait of Sicily with S. megalops from Australia (1.40%), with Squalus cf. megalops from Mauritius (6.50%) and with Squalus sp. Clade C (sensu Veríssimo et al. Citation2017), which included individuals S7 and S9 collected in the Strait (6.60%; Table S3). NADH2 genetic distances showed similar results. In particular, distances increased when comparing Squalus sp. caught in the Strait of Sicily with S. blainville (0.52%), with Squalus sp. Clade B (sensu Veríssimo et al. Citation2017; 0.56%), with Squalus cf. megalops from Mauritius (4.95%) and with Squalus sp. Clade C (sensu Veríssimo et al. Citation2017), which included S7 and S9 individuals (5%; Table S4).
The haplotype network in Figure S3 showed the presence of two most frequent haplotypes shared by almost all the samples ascribable to S. blainville. Clusters of fewer individuals sharing the same haplotype, down to one individual displaying only a different variant, are separated from one to five mutational steps. The S. blainville haplogroups were separated from Squalus samples collected in Sicily and Malta and shared haplotypes with Squalus sp. Clade C from Gabon and Guinea, by at least 68 mutations. Furthermore, 59 mutational steps separated the S. blainville haplogroups from Squalus cf. megalops (Mauritius), while the latter was separated from Squalus sp. Clade C by 24 mutations.
Discussion
The use of molecular taxonomy is largely considered a powerful tool for the correct assessment of specimens identification, the discovery of new species and, in some cases, also the identification of cryptic (Duncan et al. Citation2006; Corrigan et al. Citation2008; Liu et al. Citation2013) and/or intricate complexes of species (Ward et al. Citation2008; Arlyza et al. Citation2013). Indeed, among sharks and batoids, new species have been described (De Astarloa et al. Citation2008; Smith et al. Citation2008; White & Iglesias Citation2011; Daly-Engel et al. Citation2018; Pfleger et al. Citation2018), old species resurrected (Ebert et al. Citation2010; Viana & de Carvalho Citation2018), and identification issues have been resolved with DNA barcoding when the morphological methods gave misleading results (Bonello et al. Citation2016; Cariani et al. Citation2017).
The lack of robust data from original descriptions compromises the correct identification of specimens with implications for species conservation and management purposes. This is particularly true for many genera, Squalus included (Veríssimo et al. Citation2017). The integration of morphological and molecular taxonomy techniques has been proved helpful (Henderson et al. Citation2016). In the last years, few studies in the context of integrative taxonomy were successfully performed on the genus Squalus aiming at the integration of new molecular taxonomy techniques to more classical morphological analyses with the aim to clarify taxonomic ambiguities between some of the species (Marouani et al. Citation2012; Bonello et al. Citation2016).
The evidence obtained in the present study, which followed in the footsteps of previous case studies in terms of genetic methodologies and analytical approach (Ward et al. Citation2007; Naylor et al. Citation2012; Vella et al. Citation2017; Veríssimo et al. Citation2017; Bellodi et al. Citation2018), confirmed the branching of the genus Squalus into three main lineages as referring to Squalus acanthias/S. suckleyi (group I), S. blainville/S. megalops/S. raoulensis/S. brevirostris (group II) and a heterogeneous group of species mainly represented by the S. mitsukurii species complex (group III). At a finer resolution, the assignment of public data of Squalus individuals to Clades A-D (sensu Veríssimo et al. Citation2017) was confirmed by robust tree topologies. Almost all the individuals sampled in the Strait of Sicily and classified as Squalus sp. clustered in Clade B (sensu Veríssimo et al. Citation2017) and thus we suggest that they could be assigned to the nominal species S. blainville. On the other hand, two individuals sampled in the Strait of Sicily and classified as Squalus sp. fell in Clade C, together with four more specimens collected in the adjacent Tunisia (BOLD: FOAI329-09) and Malta waters (data from Vella et al. Citation2017) and showed a genetic distance from the former Clade B as high as the one existing between well-distinguished nominal species (e.g. S. mitsukurii and S. raoulensis from group III and II, respectively). Similar distances between genetic lineages were described in Veríssimo et al. (Citation2017).
Therefore, here we confirm the hypothesis that specimens of Clade C are not related to the Australian S. megalops, since these lineages are separated by a distance of 6.20%. In fact, individuals of Squalus sp. from Clade C are closer to S. megalops from Mauritius (2.30% according to the COI gene). This was confirmed by the species-haplotype network, where at least 68 mutational steps separated Squalus sp. clustering in Clade B (ascribable to S. blainville) from Squalus sp. clustering in Clade C from tropical Western Africa. Similar haplotype differentiation between these same lineages was observed by Vella et al. (Citation2017), who reported 83 mutations across a concatenated dataset 1600 bp long.
Hence, here we support previous studies that suggest that S. megalops does not occur in the eastern Atlantic and Mediterranean waters and that individuals composing Clade C should be considered a new species that needs formal description and proper taxonomic assessment (Last & Stevens Citation2009; Veríssimo et al. Citation2017). As a matter of fact, the occurrence of S. megalops appears more likely limited to the Australian waters, since the tree topologies obtained herein showed Squalus blainville from the Strait of Sicily (this study) clustering with longnose spurdog S. blainville from Sardinian coasts (Bellodi et al. Citation2018), Malta (Bonello et al. Citation2016; Vella et al. Citation2017), Tuscany (this study), Spain, Libyan and Ionian Seas, Greece (Kousteni et al. Citation2016) along with Squalus sp. Clade B (Veríssimo et al. Citation2017). Unfortunately, the sequence data from Marouani et al. (Citation2012) are not publicly available and thus a direct comparison was not possible.
The results discussed here, reinforced by both the genetic distances measured between groups and clades and the haplotype network, support the hypothesis that a common species, S. blainville, is currently inhabiting the Mediterranean Sea, while a second and extremely rare one, which has not yet been extensively described, is probably an occasional visitor of the Strait. This second species shows a strikingly high morphological similarity to S. blainville, but it is genetically distinct from both S. blainville and S. megalops. Within this perspective, the absence of records of S. megalops in the Atlantic Ocean and Mediterranean Sea (Straube et al. Citation2013; Veríssimo et al. Citation2017; Viana & de Carvalho Citation2018) can be confirmed. As previously mentioned, cryptic speciation among elasmobranchs is very common (Borsa et al. Citation2016, Citation2018) and the number of new descriptions, re-descriptions and resurrections of species is growing with the increasing application of molecular tools and integrated taxonomic methodologies. Starting from the accurate morphological data registered in Marouani et al. (Citation2012) a dedicated effort is needed to identify and assess diagnostic features that characterize the individuals ascribed to Clade C exhibiting high morphological similarity with both species (S. megalops and S. blainville). What is more, a comparative approach involving both mtDNA markers and highly polymorphic nuclear DNA loci, besides morphology, is recommended to enhance the power of analyses aiming at unravelling the gene flow and migratory patterns of such a rare and geographically limited species.
To conclude, current inconsistencies in species identification within the genus Squalus need to be resolved and an accurate redescription of Squalus species is advised, especially in the Mediterranean where, along with commercial fishery, the bycatch of sharks may lead to the erosion of local biodiversity including genetic diversity. For all these reasons, the growing concern about the vulnerability of sharks to fishing pressure (Dulvy et al. Citation2014) and overexploitation (Simpfendorfer & Kyne Citation2009) now requires strong measures for species protection and management, especially in this exploited stretch of Sea.
To continue the acquisition of new information and the resolution of old problems, the establishment and strengthening of an international network of collaborations and the maximisation of sampling effort would go a long way towards filling the gaps in our knowledge of these shark species, which represent an irreplaceable component of biodiversity, in terms of both species and genetic richness, to be protected and conserved before they are “Lost Before Found” (White et al. Citation2019).
Supplemental Material
Download MS Excel (70.2 KB)Supplemental Material
Download MS Excel (2.1 MB)Supplemental Material
Download MS Excel (2.6 MB)Supplemental Material
Download MS Excel (14.7 KB)Supplemental Material
Download MS Word (6.6 KB)Supplemental Material
Download JPEG Image (828.9 KB)Supplemental Material
Download TIFF Image (1.7 MB)Supplemental Material
Download TIFF Image (1.7 MB)Acknowledgements
We thank Mariano Enrico Giarrusso (University of Bologna) for his contribution in the analysis of Squalus specimens. We also thank all contributors to this research. In particular, we are grateful to the MEDITS partners for their sampling effort.
Disclosure statement
No potential conflict of interest was reported by the authors.
Supplementary material
Supplemental data for this article can be accessed here.
Additional information
Funding
References
- Arlyza IS, Shen KN, Solihin DD, Soedharma D, Berrebi P, Borsa P. 2013. Species boundaries in the Himantura uarnak species complex (Myliobatiformes: Dasyatidae). Molecular Phylogenetics and Evolution 66(1):429–435. DOI: 10.1016/j.ympev.2012.09.023.
- Aschliman NC, Nishida M, Miya M, Inoue JG, Rosana KM, Naylor GJ. 2012. Body plan convergence in the evolution of skates and rays (Chondrichthyes: Batoidea). Molecular Phylogenetics and Evolution 63(1):28–42. DOI: 10.1016/j.ympev.2011.12.012.
- Avise JC. 2004. What is the field of biogeography, and where is it going? Taxon 53(4):893–898. DOI: 10.2307/25065345.
- Bandelt H, Forster P, Röhl A. 1999. Median-joining networks for inferring intraspecific phylogenies. Molecular Biology and Evolution 16(1):37–48. DOI: 10.1093/oxfordjournals.molbev.a026036.
- Barbuto M, Galimberti A, Ferri E, Labra M, Malandra R, Galli P, Casiraghi M. 2010. DNA barcoding reveals fraudulent substitutions in shark seafood products: The Italian case of “palombo” (Mustelus spp.). Food Research International 43(1):376–381. DOI: 10.1016/j.foodres.2009.10.009.
- Bellodi A, Porcu C, Cau A, Marongiu M, Melis R, Mulas A, Pesci P, Follesa M, Cannas R. 2018. Investigation on the genus Squalus in the Sardinian waters (Central-Western Mediterranean) with implications on its management. Mediterranean Marine Science 19(2):256–272. DOI: 10.12681/mms.15426.
- Bonello J, Bonnici L, Ferrari A, Cariani A, Schembri P. 2016. Not all that clear cut: Intraspecific morphological variability in Squalus blainville (Risso, 1827) and implications for identification of the species. Journal of the Marine Biological Association of the United Kingdom 96(8):1585–1596. DOI: 10.1017/S0025315415001915.
- Borsa P, Arlyza IS, Hoareau TB, Shen KN. 2018. Diagnostic description and geographic distribution of four new cryptic species of the blue-spotted maskray species complex (Myliobatoidei: Dasyatidae; Neotrygon spp.) based on DNA sequences. Journal of Oceanology and Limnology 36(3):827–841. DOI: 10.1007/s00343-018-7056-2.
- Borsa P, Shen KN, Arlyza IS, Hoareau TB. 2016. Multiple cryptic species in the blue-spotted maskray (Myliobatoidei: Dasyatidae: Neotrygon spp.): An update. Comptes rendus biologies 339(9–10):417–426. DOI: 10.1016/j.crvi.2016.07.004.
- Cannizzaro L, Rizzo P, Levi D, Gancitano S. 1995. Age determination and growth of Squalus blainvillei (Risso, 1826). Fisheries Research 23(1–2):113–125. DOI: 10.1016/0165-7836(94)00333-R.
- Cariani A, Messinetti S, Ferrari A, Arculeo M, Bonello JJ, Bonnici L, Cannas R, Carbonara P, Cau A, Charilaou C, Ouamari NE, Fiorentino F, Follesa MC, Garofalo G, Golani D, Guarniero I, Hanner R, Hemida F, Kada O, Lo Brutto S, Mancusi S, Morey G, Schembri PJ, Serena F, Sion L, Stagioni M, Tursi A, Vrgoc N, Steinke D, Tinti F. 2017. Improving the conservation of mediterranean chondrichthyans: The ELASMOMED DNA barcode reference library. PloS One 12(1):e0170244. DOI: 10.1371/journal.pone.0170244.
- Chen H, Chen X, Yu H, Ai W. 2016. Complete mitochondrial genome and the phylogenetic position of the Taiwan spurdog shark Squalus formosus (Squaliformes: Squalidae). Mitochondrial DNA Part B 1(1):419–420. DOI: 10.1080/23802359.2016.1176884.
- Collins RA, Cruickshank RH. 2013. The seven deadly sins of DNA barcoding. Molecular Ecology Resources 13(6):969–975.
- Compagno LJV. 1984. Squaliformes-Dogfish sharks. In: Fischer W, Nauen C, editors. FAO species catalogue, Volume 4. Sharks of the world: An annotated and illustrated catalogue of shark species known to date. Part I. hexanchiformes to lamniformes. Vol. 125. Rome: FAO Fisheries Synopsis. pp. 111–113.
- Corrigan S, Huveneers C, Schwartz TS, Harcourt RG, Beheregaray LB. 2008. Genetic and reproductive evidence for two species of ornate wobbegong shark Orectolobus spp. on the Australian east coast. Journal of Fish Biology 73(7):1662–1675. DOI: 10.1111/j.1095-8649.2008.02039.x.
- Daly-Engel TS, Koch A, Anderson JM, Cotton CF, Grubbs RD. 2018. Description of a new deep-water dogfish shark from Hawaii, with comments on the Squalus mitsukurii species complex in the West Pacific. ZooKeys 798:135–157. DOI: 10.3897/zookeys.798.28375.
- De Astarloa JD, Mabragana E, Hanner R, Figueroa DE. 2008. Morphological and molecular evidence for a new species of longnose skate (Rajiformes: Rajidae: Dipturus) from Argentinean waters based on DNA barcoding. Zootaxa 1921(1):35–46. DOI: 10.11646/zootaxa.1921.1.3.
- Dulvy NK, Fowler SL, Musick JA, Cavanagh RD, Kyne PM, Harrison LR, Carlson JK, Davidson LNK, Fordham SV, Francis MP, Pollock CM, Simpfendorfer CA, Burgess GH, Carpenter KE, Compagno LJV, Ebert DA, Gibson C, Heupel MR, Livingstone SR, Sanciangco JC, Stevens JD, Valenti S, White WT. 2014. Extinction risk and conservation of the world’s sharks and rays. eLife 3:e00590.
- Duncan KM, Martin AP, Bowen BW, De Couet HG. 2006. Global phylogeography of the scalloped hammerhead shark (Sphyrna lewini). Molecular Ecology 15(8):2239. DOI: 10.1111/j.1365-294X.2006.02933.x.
- Ebert DA, Stehmann MFW. 2013. Sharks, batoids, and chimaeras of the North Atlantic. Rome: FAO.
- Ebert DA, White WT, Goldman KJ, Compagno LJ, Daly-Engel TS, Ward RD. 2010. Resurrection and redescription of Squalus suckleyi (Girard, 1854) from the North Pacific, with comments on the Squalus acanthias subgroup (Squaliformes: Squalidae). Zootaxa 2612(1):22–40. DOI: 10.11646/zootaxa.2612.1.2.
- Felsenstein J. 1985. Confidence limits on phylogenies: An approach using the bootstrap. Evolution 39(4):783–791. DOI: 10.2307/2408678.
- Garrick JAF. 1960. Studies on New Zealand Elasmobranchii. Part XII. The species of Squalus from New Zealand and Australia; and a general account and key to the New Zealand Squaloidea. Transactions of the Royal Society of New Zealand 88:519–557.
- Henderson AC, Reeve AJ, Jabado RW, Naylor GJP. 2016. Taxonomic assessment of sharks, rays and guitarfishes (Chondrichthyes: Elasmobranchii) from south-eastern Arabia, using the NADH dehydrogenase subunit 2 (NADH2) gene. Zoological Journal of the Linnean Society 176(2):399–442. DOI: 10.1111/zoj.12309.
- Hoang DT, Chernomor O, Von Haeseler A, Minh BQ, Vinh LS. 2018. UFBoot2: Improving the ultrafast bootstrap approximation. Molecular Biology and Evolution 35(2):518–522. DOI: 10.1093/molbev/msx281.
- IUCN. 2019. The IUCN red list of threatened species. Version 2019-1. Available: www.iucnredlist.org. Accessed Mar 2019 21.
- Kemper JM, Naylor GJ. 2016. The complete mitochondrial genome and phylogenetic position of the Philippines spurdog. Squalus Montalbani Mitochondrial DNA Part A 27(6):4522–4523. DOI: 10.3109/19401736.2015.1101544.
- Kousteni V, Kasapidis P, Kotoulas G, Megalofonou P. 2016. Evidence of high genetic connectivity for the longnose spurdog Squalus blainville in the Mediterranean Sea. Mediterranean Marine Science 17(2):371–383. DOI: 10.12681/mms.1222.
- Krehenwinkel H, Pomerantz A, Henderson JB, Kennedy SR, Lim JY, Swamy V, Shoobridge JD, Graham N, Patel NH, Gillespie RG, Prost S. 2019. Nanopore sequencing of long ribosomal DNA amplicons enables portable and simple biodiversity assessments with high phylogenetic resolution across broad taxonomic scale. GigaScience 8(5):giz006. DOI: 10.1093/gigascience/giz006.
- Kumar S, Stecher G, Li M, Knyaz C, Tamura K. 2018. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Molecular Biology and Evolution 35(6):1547–1549. DOI: 10.1093/molbev/msy096.
- Landi M, Dimech M, Arculeo M, Biondo G, Martins R, Carneiro M, Carvalho GR, Brutto SL, Costa FO. 2014. DNA barcoding for species assignment: The case of mediterranean marine fishes. PLoS ONE 9(9):e106135. DOI: 10.1371/journal.pone.0106135.
- Last PR, Stevens JD. 2009. Sharks and rays of Australia. Second ed. Collingwood, Australia: CSIRO Publishing.
- Last PR, White WT, Pogonosky JJ (2007). Descriptions of the new dogfishes of the Genus Squalus (Squaloidea: Squalidae). CSIRO marine and atmospheric research paper 14. Hobart: CSIRO, pp. 55–70
- Leigh JW, Bryant D. 2015. PopART: Full-feature software for haplotype network construction. Methods in Ecology and Evolution 6(9):1110–1116. DOI: 10.1111/2041-210X.12410.
- Liu SYV, Chan CLC, Lin O, Hu CS, Chen CA. 2013. DNA barcoding of shark meats identify species composition and CITES-listed species from the markets in Taiwan. PloS One 8(11):e79373. DOI: 10.1371/journal.pone.0079373.
- Mabragaña E, Díaz De Astarloa JM, Hanner R, Zhang J, González Castro M. 2011. DNA barcoding identifies argentine fishes from marine and brackish waters. PLoS ONE 6(12):e28655. DOI: 10.1371/journal.pone.0028655.
- Mandelman JW, Farrington MA. 2007. The physiological status and mortality associated with otter-trawl capture, transport, and captivity of an exploited elasmobranch, Squalus acanthias. ICES Journal of Marine Science 64(1):122–130. DOI: 10.1093/icesjms/fsl003.
- Marouani S, Chaâba R, Kadri H, Saidi B, Bouain A, Maltagliati F, Last P, Séret B, Bradai MN. 2012. Taxonomic research on Squalus megalops (Macleay, 1881) and Squalus blainvillei (Risso, 1827) (Chondrichthyes: Squalidae) in Tunisian waters (central Mediterranean Sea). Scientia Marina 76(1):97–109. DOI: 10.3989/scimar.03440.22B.
- McCusker MR, Denti D, Van Guelpen L, Kenchington E, Bentzen P. 2013. Barcoding Atlantic Canada’s commonly encountered marine fishes. Molecular Ecology Resources 13:177–188. DOI: 10.1111/1755-0998.12043.
- McEachran JD, Dunn KA. 1998. Phylogenetic analysis of skates, a morphologically conservative clade of elasmobranchs (Chondrichthyes: Rajidae). Copeia (2):271–290. DOI: 10.2307/1447424.
- Milisenda G, Vitale S, Massi D, Enea M, Gancitano V, Giusto GB, Badalucco C, Gristina M, Garofalo G, Fiorentino F. 2017. Spatio-temporal composition of discard associated with the deep water rose shrimp fisheries (Parapenaeus longirostris, Lucas 1846) in the south-central Mediterranean Sea. Mediterranean Marine Science 18(1):53–63. DOI: 10.12681/mms.1787.
- Munoz-Chapuli R, Ramos F. 1989. Morphological comparison of Squalus blainvillei and S. megalops in the Eastern Atlantic, with notes on the genus. Japanese Journal of Ichthyology 36(1):6–21. DOI: 10.1007/BF02905668.
- Naylor GJ, Caira JN, Jensen J, Rosana KA, Straube N, Lakner C. 2012. Elasmobranch phylogeny: A mitochondrial estimate based on 595 species. In: Carrier JC, Musick JA, Heithaus MR, editors. Biology of sharks and their relatives. Boca Raton: CRC press. pp. 31–56.
- Nguyen LT, Schmidt HA, Von Haeseler A, Minh BQ. 2015. IQ-TREE: A fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Molecular Biology and Evolution 32(1):268–274. DOI: 10.1093/molbev/msu300.
- Pfleger MO, Grubbs RD, Cotton CF, Daly-Engel TS. 2018. Squalus clarkae sp. nov., a new dogfish shark from the Northwest Atlantic and Gulf of Mexico, with comments on the Squalus mitsukurii species complex. Zootaxa 4444(2):101–119. DOI: 10.11646/zootaxa.4444.2.1.
- Quattro JM, Stoner DS, Driggers WB, Anderson CA, Priede KA, Hoppmann EC, Duncan KM, Grady JM. 2006. Genetic evidence of cryptic speciation within hammerhead sharks (Genus Sphyrna). Marine Biology 148(5):1143–1155. DOI: 10.1007/s00227-005-0151-x.
- Rambaut A, Drummond A (2012). FigTree: Tree figure drawing tool, v1. 4.2. Institute of Evolutionary Biology, University of Edinburgh. Available: http://tree.bio.ed.ac.uk/software/figtree
- Rasmussen A, Arnason U. 1999. Phylogenetic studies of complete mitochondrial DNA molecules place cartilaginous fishes within the tree of bony fishes. Journal of Molecular Evolution 48(1):118–123. DOI: 10.1007/PL00006439.
- Ratnasingham S, Hebert PD. 2007. BOLD: The barcode of life data system. Molecular Ecology Notes 7(3):355–364. DOI: 10.1111/j.1471-8286.2007.01678.x.
- Ronquist F, Teslenko M, van der Mark P, Ayres DL, Darling A, Höhna S, Larget B, Liu L, Suchard MA, Huelsenbeck JP. 2012. MrBayes 3.2: Efficient Bayesian phylogenetic inference and model choice across a large model space. Systematic Biology 61(3):539–542. DOI: 10.1093/sysbio/sys029.
- Rozas J, Ferrer-Mata A, Sanchez-DelBarrio JC, Guirao-Rico S, Librado P, Ramos-Onsins SE, Sanchez-Gracia A. 2017. DnaSP 6: DNA sequence polymorphism analysis of large data sets. Molecular Biology and Evolution 34(12):3299–3302. DOI: 10.1093/molbev/msx248.
- Saitou N, Nei M. 1987. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Molecular Biology and Evolution 4(4):406‐425. DOI: 10.1093/oxfordjournals.molbev.a040454.
- Sembiring A, Pertiwi NPD, Mahardini A, Wulandari R, Kurniasih EM, Kuncoro AW, Cahyani NKD, Anggoro AW, Ulfa M, Madduppa H, Carpenter KE, Barber PH, Mahardika GN. 2015. DNA barcoding reveals targeted fisheries for endangered sharks in Indonesia. Fisheries Research 164:130–134. DOI: 10.1016/j.fishres.2014.11.003.
- Serena F, Papacostantinou C, Relini G, Gil De Sola L, Bertrand J. 2009. Distribution and abundance of spiny dogfish in the mediterranean sea based on the mediterranean international trawl surveys program. In: Gallucci VF, McFarlane GA, Bargmann GC, editors. Biology and management of dogfish sharks. American fisheries society. Maryland: Bethesda. pp. 139–149.
- Simpfendorfer CA, Kyne PM. 2009. Limited potential to recover from overfishing raises concerns for deep-sea sharks, rays and chimaeras. Environmental Conservation 36(2):97–103. DOI: 10.1017/S0376892909990191.
- Smith PJ, Steinke D, Mcveagh SM, Stewart AL, Struthers CD, Roberts CD. 2008. Molecular analysis of Southern Ocean skates (Bathyraja) reveals a new species of Antarctic skate. Journal of Fish Biology 73(5):1170–1182. DOI: 10.1111/j.1095-8649.2008.01957.x.
- Smith SA, Dunn CW. 2008. Phyutility: A phyloinformatics tool for trees, alignments and molecular data. Bioinformatics 24(5):715–716. DOI: 10.1093/bioinformatics/btm619.
- Song H, Buhay JE, Whiting MF, Crandall KA. 2008. Many species in one: DNA barcoding overestimates the number of species when nuclear mitochondrial pseudogenes are coamplified. Proceedings of the National Academy of Sciences 105(36):13486–13491. DOI: 10.1073/pnas.0803076105.
- Spedicato MT, Massutí E, Mérigot B, Tserpes G, Jadaud A, Relini G. 2019. The MEDITS trawl survey specifications in an ecosystem approach to fishery management. Scientia Marina 83(S1):9–20. DOI: 10.3989/scimar.04915.11X.
- Steinke D, Zemlak TS, Gavin H, Hebert PDN. 2009. DNA barcoding fishes of the Canadian Pacific. Marine Biology 156(12):2641–2647. DOI: 10.1007/s00227-009-1284-0.
- Stoner AW, Kaimmer SM. 2008. Reducing elasmobranch bycatch: Laboratory investigation of rare earth metal and magnetic deterrents with spiny dogfish and Pacific halibut. Fisheries Research 92(2–3):162–168. DOI: 10.1016/j.fishres.2008.01.004.
- Straube N, White WT, Ho HC, Rochel E, Corrigan S, Li C, Naylor GJ. 2013. A DNA sequence-based identification checklist for Taiwanese chondrichthyans. Zootaxa 3752(1):256–278. DOI: 10.11646/zootaxa.3752.1.16.
- Tallack SML, Mandelman JW. 2009. Do rare-earth metals deter spiny dogfish? A feasibility study on the use of electropositive “mischmetal” to reduce the bycatch of Squalus acanthias by hook gear in the Gulf of Maine. ICES Journal of Marine Science 66(2):315–322. DOI: 10.1093/icesjms/fsn215.
- Thompson JD, Higgins DG, Gibson TJ. 1994. CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Research 22(22):4673–4680. DOI: 10.1093/nar/22.22.4673.
- Vella A, Vella N, Schembri S. 2017. A molecular approach towards taxonomic identification of elasmobranch species from Maltese fisheries landings. Marine Genomics 36:17–23. DOI:10.1016/j.margen.2017.08.008.
- Veríssimo A, Cotton CF, Buch RH, Guallart J, Burgess GH. 2014. Species diversity of the deep-water gulper sharks (Squaliformes: Centrophoridae: Centrophorus) in North Atlantic waters - current status and taxonomic issues. Zoological Journal of the Linnean Society 172(4):803–830. DOI: 10.1111/zoj.12194.
- Veríssimo A, McDowell JR, Graves JE. 2010. Global population structure of the spiny dogfish Squalus acanthias, a temperate shark with an antitropical distribution. Molecular Ecology 19(8):1651–1662. DOI: 10.1111/j.1365-294X.2010.04598.x.
- Veríssimo A, Zaera-Perez D, Leslie R, Iglesias SP, Seter B, Grigoriou P, Sterioti A, Gubili C, Barria C, Duffy C, Hernandez S, Batjakas IE, Griffiths AM. 2017. Molecular diversity and distribution of eastern Atlantic and Mediterranean dogfishes Squalus highlight taxonomic issue in the genus. Zoologica scripta 46(4):414–428. DOI: 10.1111/zsc.12224.
- Viana ST, de Carvalho MR. 2018. Resurrection and redescription of the Southern Dogfish Squalus probatovi (Squalidae), a valid species from Angola. Journal of Ichtyology 58(5):617–632. DOI: 10.1134/S003294521805020X.
- Viana ST, de Carvalho MR, Gomes UL. 2016. Taxonomy and morphology of species of the genus Squalus Linnaeus, 1758 from the Southwestern Atlantic Ocean (Chondrichthyes: Squaliformes: Squalidae). Zootaxa 4133(1):1–89. DOI: 10.11646/zootaxa.4133.1.1.
- Ward RD, Holmes BH, Zemlak TS, Smith PJ. 2007. DNA barcoding discriminates spurdogs of the genus Squalus. In: Last PR, White WT, Pogonoski JJ, editors. Descriptions of new dogfishes of new dogfishes of the Genus Squalus (Squaloidea: Squalidae). CSIRO marine and atmospheric research paper 014. Hobart: CSIRO. pp. 117–130.
- Ward RD, Holmes BH, White WT, Last PR. 2008. DNA barcoding Australasian chondrichthyans: Results and potential uses in conservation. Marine & Freshwater Research 59(1):57–71. DOI: 10.1071/MF07148.
- Ward RD, Zemlak TS, Innes BH, Last PR, Hebert PDN. 2005. DNA barcoding Australia’s fish species. Philosophical Transactions of the Royal Society B 360(1462):1847–1857. DOI: 10.1098/rstb.2005.1716.
- White WT, Iglesias SP. 2011. Squalus formosus, a new species of spurdog shark (Squaliformes: Squalidae), from the western North Pacific Ocean. Journal of Fish Biology 79(4):954–968. DOI: 10.1111/j.1095-8649.2011.03068.x.
- White WT, Kyne PM, Harris M. 2019. Lost before found: A new species of whaler shark Charcarhinus obsolerus from the Western central pacific known only from historic records. PloS One 14(1):e0209387. DOI: 10.1371/journal.pone.0209387.
- Whitehead PJP, Bauchot ML, Hureau JC, Nielsen J, Tortonese E. 1984. Fishes of the North-Eastern atlantic and the mediterranean (FNAM), volumes 1–3. Paris: Unesco. pp. 1473.
- Wong EHK, Shivji MS, Robert H, Hanner RH 2009. Identifying sharks with DNA barcodes: Assessing the utility of a nucleotide diagnostic approach. Molecular Ecology Resources 9(S1):243–256. DOI: 10.1111/j.1755-0998.2009.02653.x.
- Zhang N, Guo HY, Guo L, Zhu KC, Liu BS, Yang JW, Zhang DC. 2019. Characterization of the complete mitochondrial genome of Squalus brevirostris (Squaliformes, Squalidae). Mitochondrial DNA Part B 4(2):2902–2903.
