Abstract
In the present paper, we examined the type material as well as newly collected specimens of the rare Antarctic eutardigrade species – Dastychius improvisus. Based on the available material, we prepared a full re-description of this species emending its description with new morphometrical characters. We conducted also phylogenetic analyses based on 18S and 28S rRNA sequence data and showed the position of the genus Dastychius in the family Isohypsibiidae. Moreover, we indicated that phylogenetic relationships within the superfamily Isohypsibioidea are still unresolved. This problem is mainly related to the relatively limited number of available sequences for the species from Isohypsibioidea and incorrectly identified sequences deposited in genetic databases, e.g. from the genus Eremobiotus. Finally, we also discussed the status of several members of Isohypsibioidea.
Introduction
Antarctica is the most extreme continent on Earth, with very low temperatures and the near absence of liquid water. Among ca. 14 million square kilometres of its area, only ca. 0.3% is ice-free. The continent is divided into three main terrestrial units: i.e. East Antarctica, West Antarctica and the Antarctic Peninsula (which is characterized by the richest biodiversity in this region (Riffenburh Citation2007)). The Antarctic also can be divided into continental part and maritime part. The last have the most rich biodiversity in the region and include Alexander Island, the archipelagos of the South Shetland, South Orkney, South Sandwich Islands, and the isolated Bouvet and Peter I Islands (Kimble Citation2004). Antarctic Peninsula has the most diverse vegetation (algae, lichens, liverworts, mosses and two species of vascular plants) in the Antarctic (Lindsay Citation1971; Smith & Corner Citation1973). Our knowledge on microinvertebrates in the Antarctic is patchy because most studies are focused on well-accessible regions close to research stations (Velasco-Castrillón et al. Citation2014). Many of Antarctic invertebrates are endemic, some of them have a continental distribution, others are restricted only to maritime Antarctic and only a few can be characterized as pan-Antarctic species (e.g. Pugh & Convey Citation2008; Cesari et al. Citation2016a).
The phylum Tardigrada (water bears) currently consists of more than 1,300 species (Guidetti & Bertolani Citation2005; Degma & Guidetti Citation2007; Vicente & Bertolani Citation2013; Degma et al. Citation2020) that inhabit terrestrial and aquatic environments throughout the world. They are small invertebrates between 50 and 1200 µm in length. The body of tardigrades is divided into five segments (head and four trunk segments with legs). The phylum Tardigrada is divided into two main classes, i.e. Eutardigrada and Heterotardigrada. Among the Eutardigrada there are species with soft and elastic cuticle (mainly terrestrial and freshwater species), but in most of Heterotardigrada dorsal side of the body is covered by cuticular plates (terrestrial and marine species). As for internal organs, the tardigrades have reproductive, excretory, digestive, muscular and nervous systems, but the respiratory and circulatory systems are absent. Tardigrades feed mainly on moss cells, algae, rotifers, nematodes, Bacteria and other tardigrades. The most extraordinary ability of tardigrades is cryptobiosis. It is a dormant state in which water bears are able to withstand extreme environmental conditions, e.g. lack of water and very high and very low temperatures. Due to their ability to cryptobiosis, they are able to inhabit and survive in extreme environments that are hostile for almost all other invertebrates such as polar deserts in Antarctica (Ramazzotti & Maucci Citation1983; Nelson et al. Citation2015).
Studies on Antarctic tardigrades started at the beginning of the twentieth century and have progressed slowly up to the present. Currently, 71 tardigrades have been reported from the continental Antarctic and the Antarctic Peninsula, and a few from other regions (for review, see Convey & McInnes Citation2005; and later publications:; Binda et al. Citation2005; McInnes Citation2010; Pilato et al. Citation2012; Guidetti et al. Citation2014; Kaczmarek et al. Citation2014; Tsujimoto et al. Citation2014; Velasco-Castrillón et al. Citation2014; Vecchi et al. Citation2016; Guidetti et al. Citation2017; Pilato et al. Citation2017; Dastych Citation2018; Kaczmarek et al. Citation2018, Citation2020a; Kihm et al. Citation2020; Tsujimoto et al. Citation2020).
Dastychius improvisus (Dastych, Citation1984) was described from Enderby Land and King George Island by Dastych (Citation1984) as Isohypsibius improvisus. Later, mainly based on shape of apophyses for the insertion of stylet muscles (AISM), it was attributed to the newly described genus Dastychius Pilato, Citation2013 and the family Isohypsibiidae Sands, McInnes, Marley, Goodall-Copestake, Convey and Linse, 2008 (Sands et al. Citation2008; Pilato Citation2013). Recently, Guil et al. (Citation2019) revised the entire taxonomy of Tardigrada and proposed a number of new taxonomic ranks, e.g. new orders which replaced superfamilies, but this revision was just recently partially questioned in case of Heterotardigrada and Apochela by Gąsiorek and Michalczyk (Citation2020) and Morek et al. (Citation2020). Another two genera, i.e. Vladimirobius and Weglarskobius in that superfamily were described by Kaczmarek et al. (Citation2020b). According to the currently accepted phylogeny, the genus Dastychius belongs to the family Isohypsibiidae and superfamily Isohypsibioidea Sands, McInnes, Marley, Goodall-Copestake, Convey and Linse, 2008. The genus is monotypic and endemic for Antarctic region. Isohypsibioidea is still one of the major tardigrade groups with unresolved systematics and phylogenetic relationships (Guil et al. Citation2019). Insufficient sampling of Isohypsibioidea resulted in predominant polytomies of its phylogeny (Bertolani et al. Citation2014a; Gąsiorek et al. Citation2019a) and only one study indicated a monophyletic lineage within this superfamily of tardigrades (Cesari et al. Citation2016b). Nevertheless, Gąsiorek et al. (Citation2019b) performed recently a broad taxon sampling and reported subclades with unresolved phylogenetic relationships, distinct phyletic lineages in paraphyletic relationships as well as monophyletic relationships within Isohypsibioidea.
In the present study, we examined paratypes (from Zoologisches Museum of Hamburg, Germany) of Das. improvisus and fresh material of this species from the Vechernij region of the Tala Hills oasis, Enderby Land. Based on paratypes and newly collected material, we prepared a full set of measurements and microphotographs of this species and described some new morphological structures reported neither in the original description nor later in partial re-description by Pilato (Citation2013). On the basis of molecular analyses of 18S rRNA and 28S rRNA sequences, we confirmed the phylogenetic position of the genus Dastychius in the family Isohypsibiidae. Moreover, we indicated that phylogenetic relationships within the superfamily Isohypsibioidea are still unresolved. Finally, we also discuss the status of a few members of Isohypsibioidea, i.e. Doryphoribius flavus (Iharos, Citation1966), Doryphoribius macrodon (Iharos, Citation1966), Thulinius augusti (Murray, Citation1907), Fractonotus verrucosus (Richters, Citation1900), Eremobiotus alicatai (Binda, Citation1969) and Isohypsibius dastychi Pilato, Bertolani & Binda, Citation1982.
Materials and methods
Sampling
A lichen (Umbilicaria aprina Nyl., 1863) sample was collected from the surroundings of Belarusian Antarctic Station Vechernyaya Mount (67°39′ S, 46°09′ E) of the Tala Hills Oasis, East Antarctica during seasonal work of the ninth Belarusian Antarctic expedition (2016–2017). The dried sample was packed in a sterile bottle (100 ml) in the field and delivered to the laboratory at the Faculty of Biology, Adam Mickiewicz University, Poznań, Poland. Tardigrades were extracted from the sample according to the method described in Stec et al. (Citation2015).
The State Research and Production Association “Scientific and Practical Center of the National Academy of Sciences of Belarus for Bioresources” (abbreviated as SPC for Bioresources) gave the permissions to conduct the sampling in surroundings of the Belarusian Antarctic station during seasonal Belarusian Antarctic Expeditions (abbreviated as BAE). The SPC for Bioresources is a part of the National Academy of Sciences of Belarus (the state competent authority of the National Antarctic Program of Belarus “Monitoring of the Earth’s Polar Areas, construction of Belarusian Antarctic Station and Promotion of Polar Expeditions”) and it is responsible for biodiversity studies in Antarctica.
Microscopy and imaging
Specimens for light microscopy were mounted on microscope slides in a small drop of Hoyer’s medium, prepared according to Ramazzotti and Maucci (Citation1983) as in the English translation by Beasley (Citation1995), and secured with a cover slip. The slides were then placed in an incubator and dried for 2 days at ca. 60°C. Dried slides were sealed with a transparent nail polish and examined under an Olympus BX41 phase contrast light microscope (PCM) associated with an ARTCAM–300Mi digital camera (Olympus Corporation, Shinjuku–ku, Japan).
All figures were assembled in Corel Photo-Paint 2017. For deep structures that could not be fully focused in a single photograph, a series of 2–10 images were taken every ca. 0.5 μm and then manually assembled into a single deep-focus image in Corel Photo-Paint 2017.
Morphometrics and morphological nomenclature
All measurements, taken using the QuickPhoto Camera 2.3 software, are given in micrometres (μm). Structures were measured only if their orientation was suitable. Body length was measured from the anterior extremity to the end of the body, excluding the hind legs. The types of bucco-pharyngeal apparatuses and claws were classified according to Pilato and Binda (Citation2010) and Pilato (Citation2013). The buccal tube was measured according to Kaczmarek and Michalczyk (Citation2017). Macroplacoid length sequence is presented according to Kaczmarek et al. (Citation2014). Lengths of the claw branches were measured according to Beasley et al. (Citation2008). The pt ratio is the ratio of the length of a given structure to the length of the buccal tube, expressed as a percentage (Pilato Citation1981). The pt values are always provided in italics.
Morphometric data were handled using the “Parachela” ver. 1.6 template available from the Tardigrada Register (Michalczyk & Kaczmarek Citation2013). Tardigrade taxonomy follows Bertolani et al. (Citation2014a). Genus abbreviations follow Perry et al. (Citation2019).
Comparative material
Species were identified based on the original description (Dastych Citation1984) as well as the later partial re-description (Pilato Citation2013). Additionally, the type material (four paratypes) from the collection of Zoologisches Museum of Hamburg (Germany) was also examined ().
DNA extraction, PCR amplification, molecular marker sequencing and phylogenetic analysis
Prior to DNA extraction, five specimens were preliminarily identified in vivo under PCM (400x magnification). Subsequently, each specimen (marked as AT2.1 – AT2.5) was placed separately in a 1.5 ml Eppendorf microcentrifuge tube in 20 µl of sterile MQ H2O and kept frozen at −80°C. Genomic DNA isolation was performed according to the Chelex®100 resin (Bio-Rad) extraction method (Casquet et al. Citation2012) modified in order to obtain tardigrade exoskeletons, according to Kaczmarek et al. (Citation2019). Digestion of specimens was performed in 40 µl of 10% Chelex®100 resin solution and 0.02 mg of Proteinase K (A&A Biotechnology) for 5 h at 55°C, followed by incubation at 70°C for 15 min. Additionally, the tubes were continuously shaken (500 RPM, Eppendorf Thermomixer compact 5350) and centrifuged every 30 minutes (Kaczmarek et al. Citation2019). Finally, 20 µl of sterile MQ H2O was added to each probe and they were centrifuged for 2 min at 8066 RPM. In total, 40 µl of DNA extract was carefully transferred for further analyses and the remaining extract with tardigrade exoskeleton was examined under stereomicroscope in order to remove the exoskeleton, which was later fixed on a microscope slide in Hoyer’s medium ()) and deposited in the collection of the Department of Animal Taxonomy and Ecology, Faculty of Biology, Adam Mickiewicz University, Poznań.
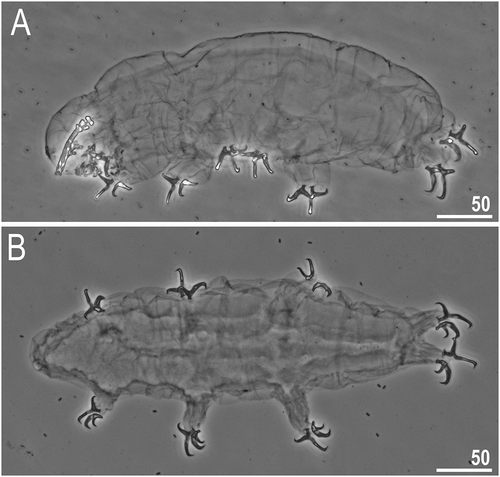
Figure 2. Dastychius improvisus, (additional material, exoskeletons): (a) lateral projection of the entire animal with bucco-pharyngeal apparatus (first exoskeleton); (b) dorso-ventral projection of the entire animal (second exoskeleton). All PCM. Scale bars in µm
Genomic regions comprising genes encoding nuclear ribosomal 18S and 28S subunits were used to assess the phylogenetic position of the Das. improvisus. We used SSU01_F (5ʹ-AACCTGGTTGATCCTGCCAGT-3ʹ) and SSU82_R (5ʹ-TGATCCTTCTGCAGGTTCACCTAC-3ʹ) primers to amplify the 18S gene fragment (Sands et al. Citation2008). The 28S rRNA gene fragment was amplified using 28SF0001 (5ʹ-ACCCvCynAATTTAAGCATAT-3ʹ) and 28SR0990 (5ʹ-CCTTGGTCCGTGTTTCAAGAC-3ʹ) primers (Miranov et al. Citation2012). The COI gene fragment was amplified using universal primers: LCO1490 (5ʹ-GGTCAACAAATCATAAAGATATTGG-3ʹ) and HCO2198 (5ʹ-TAAACTTCAGGGTGACCAAAAAATCA-3ʹ) (Folmer et al. Citation1994). The PCR reactions were performed in 20 μl volume containing 0.8× JumpStart Taq ReadyMix (1 U of JumpStart Taq DNA polymerase, 4 mM Tris–HCl, 20 mM KCl, 0.6 mM MgCl2, 0.08 mM of dNTP; Sigma-Aldrich, Germany), 0.4 μM of given forward and reverse primers and ca. 1 ng of DNA. The COI gene fragment was amplified under conditions as follows: initial denaturation at 94°C for 5 min followed by 30 cycles of 94°C for 30 s, 45°C for 1 min, and 72°C for 1 min and ending with 72°C for 5 min. The PCR cycling profile to amplify the18S rRNA gene fragment was as follows: initial denaturation at 95°C for 5 min followed by 35 cycles of 95°C for 30 s, 60°C for 1 min, and 72°C for 1 min and ending with 72°C for 5 min. In turn, PCR protocols for the amplification of the 28S rRNA gene fragment were described in Mironov et al. (Citation2012). The reactions were conducted in a BiometraTProfessional thermocycler. Amplification products were separated in a 1% agarose gel electrophoresis in a 1× SB buffer and visualized with Midori Green Advance DNA Stain (Genetics) under UV light. All PCR products were treated with alkaline phosphatase and exonuclease I (Thermo Scientific, Waltham, MA, USA) according to the manufacturer’s guidelines and sequenced directly in both directions using the BigDyeTM terminator cycle sequencing method.
BLAST searches (Altschul et al. Citation1990) were performed to verify the identity and homology of the obtained mitochondrial and nuclear gene fragments with sequences deposited in the NCBI database. All sequences were checked for quality and trimmed to the same length in BioEdit v. 7.2.5 (Hall Citation1999). The COI gene sequences could be unambiguously aligned without inserting gaps. However, preliminary alignment was performed in Geneious Prime v. 2019.0.4 (https://www.geneious.com) for 18S and 28S rRNA gene fragments using the following default parameters: gap opening = 15, gap extension = 6.6, DNA transition weight = 0.5 and delay divergent sequences = 30%, the alignment was then manually inspected. This program was also applied to concatenated 18S and 28S rRNA sequences. In addition, nucleotide sequences of the COI gene fragments were translated into amino acid sequences using the EMBOSS-TRANSEQ application (Rice et al. Citation2000; Goujon et al. Citation2010). The translation was successfully carried out with the invertebrate mitochondrial codon table and the −second reading frame. Haplotypes were retrieved using DNASP v.5.10.01 software (Librado et al. Citation2009). All obtained sequences have been deposited in GenBank under accession numbers: COI: MK765502-MK765506, 18S rRNA: MK737028-MK737032, 28S rRNA: MK737035-MK737039 (see also Table A1).
We estimated the phylogenetic relationship among 74 tardigrade taxa representing eight families and the apochelan Milnesium variefidum Morek, Gąsiorek, Stec, Blagden & Michalczyk, Citation2016a and Milnesium berladnicorum Ciobanu, Zawierucha, Moglan & Kaczmarek, Citation2014 sequences as outgroups to obtain the most reliable evolutionary tree, focused on the combined 18S and 28S rRNA dataset (for details see Table A1). However, only available GenBank sequences that coincided with nrDNA fragments analyzed in the present study were applied. The most appropriate model of sequence evolution was determined using jModelTest v. 2.1.4 (Darriba et al. Citation2012) with the assumptions of both the Akaike Information Criterion (AIC) and Bayesian Inference Criterion (BIC) (Posada & Buckley Citation2004). The GTR + G + I (Time Reversible model with gamma-distributed rate heterogeneity and proportion of invariable unchanging sites) was chosen as the best fitting evolution model for 18S and 28S rRNA, and was applied for the concatenated nrDNA data. Phylogenetic trees were computed by applying the Maximum Likelihood (ML) analysis. The ML trees were calculated using Mega X (Kumar et al. Citation2018) under the general settings of selected models with 1000 bootstraps. This program was also applied to calculate nucleotide sequence composition. Bayesian inference (BI) was conducted using the program MrBayes 3 (Ronquist & Huelsenbeck Citation2003). Random starting trees were used and each of four Metropolis-coupled Markov chain Monte Carlo (MCMC) were launched for 40 × 106 generations. Sampling the Markov chain was carried out in every 1000 generations. The program Tracer v1.3 (Rambaut et al. Citation2014) was used to verify the convergence of the MCMC runs and determine the correct “burn-in” for the analysis. The less 5,000 likely trees were discarded as burn-in. Final phylogenetic trees were edited in FigTree v. 1.4.2 software (http://tree.bio.ed.ac.uk/software/figtree). Calculation of evolutionary divergence between the combined 18S rRNA and 28S rRNA data sets based on uncorrected p-distances was performed using MEGA X. Uncorrected pairwise distances are provided as supplementary materials (Table A2).
Results and discussion
In the current re-description of Das. improvisus, we presented new detailed measurements of type specimens and additional material collected in a new locality. We characterized the buccal apparatus and the cuticle, and added taxonomically important information on these structures. We also presented the first DNA data for the genus Dastychius.
Taxonomic account
Phylum: Tardigrada, Doyère, Citation1840Class: Eutardigrada Richters, Citation1926Order: Parachela Schuster et al., Citation1980Superfamily: Isohypsibioidea Sands, McInnes, Marley, Goodall-Copestake, Convey and Linse, Citation2008Family: Isohypsibiidae Sands, McInnes, Marley, Goodall-Copestake, Convey and Linse, Citation2008Genus: Dastychius Pilato, Citation2013
Re-description
Dastychius improvisus (Dastych, Citation1984)(, )
Table I. Measurements and pt values of selected morphological structures of Dastychius improvisus (Dastych, Citation1984) (type material)
Table II. Measurements and pt values of selected morphological structures of Dastychius improvisus (Dastych, Citation1984) (additional material) mounted in Hoyer’s medium
Figure 3. Dastychius improvisus (additional material): (a) dorso-ventral projection of the entire animal; (b)bucco-pharyngeal apparatus (dorso-ventral projection); arrow indicates clearly narrowed terminal part of buccal tube; insert indicates peribuccal papulae probably present; (c) apophysis for the insertion of the stylet muscles, arrowhead. All PCM. Scale bars in µm
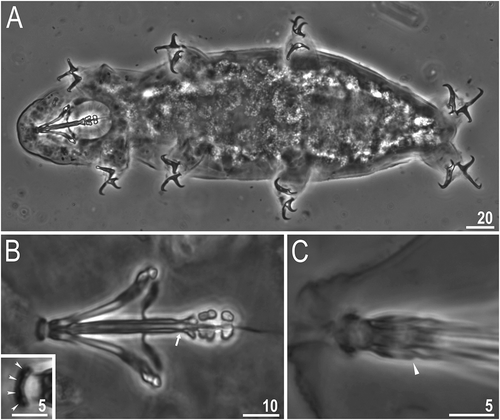
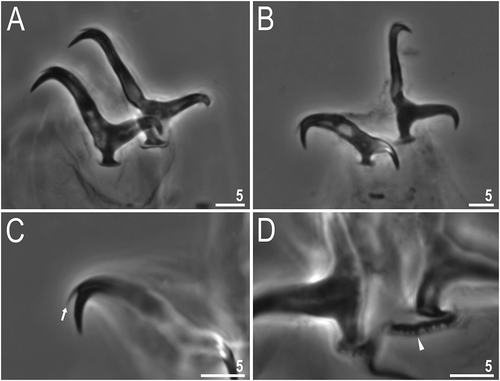
Figure 4. Dastychius improvisus (additional material): (a)claws II; (b) claws IV; (c) accessory points on claw IV; (d) dentate lunules on legs IV, arrowhead. All PCM. Scale bars in µm
Type locality
67°39′ S, 46°06′ E; East Antarctica, Enderby Land, January 1974, leg. K. Jażdżewski.
Material studied
Type material: Ant. 1726 (one paratype of Das. improvisus + Mesobiotus blocki (Dastych, Citation1984), Barbaria pseudowendti (Dastych, Citation1984) and Hebesuncus schusteri (Dastych, Citation1984)), East Antarctica, Enderby Land, Jan. 1974, 67°39′ S, 46°06′ E, lg. K. Jażdżewski; Ant. 1776 (one exuviae of Das. improvisus), East Antarctica, Enderby Land, Jan. 1974, 67°39′ S, 46°06′ E, lg. K. Jażdżewski; Ant. 34 (one paratype of Das. improvisus + Meb. blocki), East Antarctica, Enderby Land, Jan. 1974, 67°39′ S, 46°06′ E, lg. K. Jażdżewski; Ant. 1836 (one paratype of Das. improvisus + Meb. blocki and Heb. schusteri), East Antarctica, Enderby Land, Jan. 1974, 67°39′ S, 46°06′ E, lg. K. Jażdżewski. Additional material: 67°39′ S, 46°10′ E; East Antarctica, Enderby Land, Jan. 2017, lichen (Umbilicaria aprina), leg. Y. Giginiak, slides: AT2.6 (one specimen of Das. improvisus + one specimen of Hebesuncus sp.), AT2.8 (two Das. improvisus + two Hebesuncus sp. and one undefined exuviae), AT2.9 (one Das. improvisus + one Hebesuncus sp.), AT2.4 (one Das. improvisus + two Hebesuncus sp.), AT2.3 (two Das. improvisus), AT2.15 (two Das. improvisus, one Meb. blocki, nine Hebesuncus sp., three undefined exuviae), AT2.3/S (one exoskeleton Das. improvisus), AT2.4/S (one exoskeleton Das. improvisus).
Short description (measurements and statistics in )
Animals
Body in living specimens white, white-green, or sometimes with brownish pigment; transparent after preparation ()). The green coloration is caused by food is intestine (which suggests that species is herbivorous). Eyes in general present and very large (eyes were not detected in 2 of 13 examined specimens).
Dorsal and ventral cuticle smooth (i.e., without sculpturing or pores) (for more details see Remarks below). Gibbosities or other cuticular structures absent ()).
Bucco-pharyngeal apparatus of the Dastychius type ()), without ventral lamina and with apophyses for the insertion of the stylet muscles (AISM) in shape of long, continuous ridges tailing off caudally and almost reaching the level of the stylet supports (), arrowhead). Peribuccal lamellae absent but six peribuccal papulae probably present (), insert) (for details see Remarks below). Oral cavity armature absent or not visible in LM. The walls of the buccal tube (in all examined specimens) clearly narrowed in terminal part of buccal tube, directly before apophyses (), arrow). Pharyngeal bulb with triangular apophyses and with two macroplacoids. First macroplacoid in shape of short rod, second in shape of elongated granule. Macroplacoid length sequence 2 < 1. Microplacoid and septulum absent. Evident incisions present in the middle of first macroplacoid and in posterior part of second macroplacoid.
Claws of the Isohypsibius type, slightly different in shape and size on all legs ()). All primary branches with minute accessory points () arrow). Lunules present and smooth on legs I–III, and indented on IV (), arrowhead). Bars and other cuticular structures on legs absent.
Eggs
Smooth and deposited in exuviae (from 2 to 8).
Original measurements according to Dastych (Citation1984)
Body length: 360–520 μm; eyes diameter: up to 10 μm; cuticular pores: 2–7 μm (mostly 4–5 μm); buccal tube length: 54 μm (in specimen 480 μm in length); external and internal buccal tube width: 5 and 3 μm, respectively (in specimen 480 μm in length); pharynx dimension: 39 × 38 μm (in specimen 480 μm in length); placoids length: I – 6 μm, II – 4.5 μm (in specimen 480 μm in length); placoids width: 2.5 μm (in specimen 480 μm in length); claws IV: 36 μm (in specimen 480 μm in length); egg dimensions: 105–80 × 78–60 μm.
Remarks
Dastych (Citation1984) indicated that cuticle in this species is covered by numerous “cavities” which are similar to those present in the genus Macrobiotus C.A.S. Schultze, Citation1834. However, in paratypes and newly collected specimens, we only observed muscle attachments which could be interpreted as “cavities”. In our opinion the true pores, which are often present in the genus Macrobiotus, are absent in the genus Dastychius; however, this observation should be confirmed based on SEM analyses.
Dastych (Citation1984) suggested the presence of lamellae (but with doubts indicated by a question mark). Later, Pilato (Citation2013) interpreted these structures as “ … in the form of peribuccal papulae … ” and we agree with this interpretation, that these structures are probably papulae, in the number of six or ten, but this needs to be confirmed by SEM analyses.
As stated above, the walls of the buccal tube are clearly narrowed in the terminal part of buccal tube. This character was not described by Dastych (Citation1984) nor reported by Pilato (Citation2013), however is quite evident in Fig. 18d in Dastych (Citation1984). At present, we consider this character as typical for the genus Dastychius and we emend the diagnosis of the genus.
Type depositories
According to Pilato (Citation2013), the holotype* and paratypes are deposited in Binda and Pilato collection (Department of the Animal Biology, University of Catania, Italy), Zoologisches Museum of Hamburg (Germany), British Antarctic Survey Data Resource Collection (UK), National Museum of Natural History, Washington (USA) and National Museum of New Zealand, Wellington (New Zealand).
Additional specimens collected in the Vechernij region of the Tala Hills oasis Enderby Land are deposited at the Department of Animal Taxonomy and Ecology, Institute of Environmental Biology, Adam Mickiewicz University, Poznań, Uniwersytetu Poznańskiego 6, 61–614 Poznań (Poland).
*Although, Pilato (Citation2013) stated that the holotype is deposited at the Department of Animal Morphology, Adam Mickiewicz University, Poznań (Poland), this specimen is now deposited at Zoologisches Museum of Hamburg (Germany).
Genotypic differential diagnosis and phylogenetic analysis
The molecular data allow us to locate the genus Dastychius within the superfamily Isohypsibioidea and in the family Isohypsibiidae, which was suggested also by Pilato (Citation2013) based on morphological data only. Moreover, Pilato (Citation2013) stated that the most similar and closely related taxon is Ramajendas Pilato and Binda, 1990 with a very similar shape of the AISM. However, Bertolani et al. (Citation2014a), based on the shape of claws, attributed these two genera to different families, i.e. Isohypsibiidae (the genus Dastychius) and Ramazzottiidae (the genus Ramajendas). Recently, Zawierucha et al. (Citation2018) questioned the attribution of the genus Ramajendas to the family Ramazzottiidae and suggested that claws in this genus are similar to those in the genus Ramazzottius Binda and Pilato, Citation1986, but evolved independently, and the genus Ramajendas should be temporarily attributed to the family Isohypsibiidae. We agree with this statement taking into consideration the similar shape of AISM in Ramajendas and Dastychius and the fact that the molecular data confirmed attribution of the genus Dastychius to the family Isohypsibiidae.
18S rRNA and 28S rRNA
No genetic diversity in either 18S rRNA (597 bp) and 28S rRNA (806 bp) nrDNA molecular markers was observed for all five investigated Das. improvisus specimens.
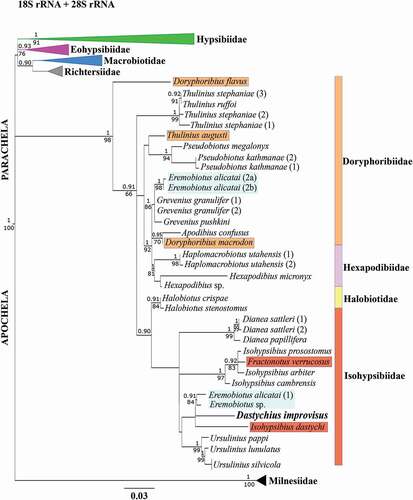
The phylogenetic position of Das. improvisus was determined based on relationships with related genera. Until now, there have been no molecular data on the genus Dastychius. What is more, nuclear sequences for species belonging to related genera are limited or different fragments of nrDNA markers are deposited since various primers were applied for the amplification. As a result, not all taxa were represented in all data sets (see Table A1). Despite the fact that the deposited 18S or 28S rRNA sequences originated from different specimen’s vouchers, and there were many species for which only one nuclear sequence was available, we performed concatenated analysis of both nrDNA markers ().
Figure 5. The phylogenetic position (Bayesian and maximum likelihood analyses) of Dastychius improvisus analyzed by the combined 18S rRNA and 28S rRNA data sets under GTR + G + I model. The supporting bootsrap values are given under branches (maximum likelihood analysis, ML); while values above branches indicate posterior probability values (Bayesian analysis, BI). Branches with support below 66% in ML and below 0.9 in BI were collapsed. The GenBank accession numbers of all the sequences applied are given in Table A1. Species with a questionable position on the phylogenetic tree were marked with appropriate colors corresponding to their family. In turn, all sequences of Eremobiotus alicatai and Eremobiotus sp. (marked in blue) are still questionable and needed revision which of them are correct
The value of uncorrected genetic p-distances between Das. improvisus and the most similar taxa, i.e. Eremobiotus alicatai (GenBank accession number: HQ604951 – 18SrRNA), Eremobiotus sp. (GenBank accession numbers: MK675928 – 18SrRNA and MK675917 – 28SrRNA) is 3.5% (see Table A2).
The other sequences of Ere. alicatai available (GenBank accession numbers: FJ435722 (18SrRNA) and FJ435766-67 (28SrRNA)) were probably incorrectly identified and may represent undefined Grevenius species. The ranges of uncorrected genetic p-distances were 0.2% between the most similar species of Gre. granulifer (Thulin, Citation1928) (GenBank accession number: EF620403 (18SrRNA)) and 5.1% between other sequences of Ere. alicatai (GenBank accession number: HQ604951 (18SrRNA)) and Eremobiotus sp. (GenBank accession numbers: MK675928 (18SrRNA) and MK675917 (28SrRNA)). Similarly, Thulinius augusti (GenBank accession number: KF360230 (18SrRNA)) is genetically most similar to Gre. granulifer with uncorrected pairwise distance value only 2.0%; however, average p-distance between Thu. stephaniae (Pilato, Citation1974) (GenBank accession numbers: GQ925701, GQ925699, AF056023 (18SrRNA)) and Thu. ruffoi (Bertolani, Citation1982) is 3.4% (GenBank accession numbers: MK675932 (18SrRNA) and MK675921 (28SrRNA)). In turn, sequence of the 18SrRNA described as Iso. dastychi (GenBank accession number: HQ604954 (18SrRNA)) is most similar to the species Ere. alicatai – 3.1% (GenBank accession numbers: HQ604951 (18SrRNA)) and the p-distance between Iso. cambrensis (Morgan, Citation1976) (the most similar of the Isohypsibius species) is 4.5% (GenBank accession number: AM500652 (18SrRNA)). The next questionable sequences belonged to Dor. macrodon (GenBank accession number: HQ604942 (18SrRNA)) and Dor. flavus (GenBank accession number: HQ604940 (18SrRNA)) - the p-distance between these species was 3.9%. The most similar species to the Dor. macrodon was Apodibius confusus Dastych, Citation1983 (GenBank accession number: KC582830 (18SrRNA) and KC582834 (28SrRNA)) – the p-distance was 1.0%. In turn, to the Dor. flavus the most similar were: Gre. granulifer (GenBank accession number: EF620403 (18SrRNA)), Gre. granulifer (GenBank accession number: KT778603 (18SrRNA)) and Gre. pushkini (Tumanov, Citation2003) (GenBank accession numbers: MK675929 (18SrRNA) and MK675918 (28SrRNA)) and the p-distance was 3.7%. Surprisingly, Fra. verrucosus (GenBank accession numbers: MG800855 (18SrRNA) and MG800856 (28SrRNA)) was clustered in subclades with Iso. prosostomus Thulin, Citation1928 (GenBank accession numbers: EF620404 (18SrRNA)), Iso. arbiter Binda, Citation1980 (GenBank accession numbers: KT778602 (18SrRNA)), Iso. cambrensis (GenBank accession numbers: AM500652 (18SrRNA)) with uncorrected pairwise distance value only from 0.6% to 1.4% (see Table A2).
COI
The length of the COI gene was 617 base pairs (bp). All sequences represented the same haplotype and contained no insertions, deletions or stop codons. Homology comparison of the very first time obtained Das. improvisus COI sequences with GenBank records (Wed, 19 Dec 2018 17:00:00 EST) indicated similarity to the phylum Tardigrada. The most closely related sequence derived from the family Isohypsibiidae, i.e. Iso. pushkini, and was in 79% identical (query coverage was 99% and E-value was 6e-113).
Phylogenetic relationships of Isohypsibioidea
Based on our analyses of 18S rRNA and 28S rRNA, we can clearly state that the phylogenetic relationships of the superfamily Isohypsibioidea (especially families Isohypsibiidae and Doryphoribiidae Gąsiorek et al. Citation2019b) are still unclear. Overall, we can confirm that the genus Dastychius belongs to the family Isohypsibiidae and that the evolutionary relationships within the family are far from being resolved, especially in case of the genus Isohypsibius Thulin, Citation1928. A similar phylogenetic reconstruction was previously conducted by Gąsiorek et al. (Citation2019b). As stated above, the phylogenetic position of some species based on sequences deposited in GenBank, i.e. Ere. alicatai (Guil & Giribet Citation2012; Bertolani et al. Citation2014a), Eremobiotus sp. (Gąsiorek et al. Citation2019b), Thu. augusti (Bertolani et al. Citation2014a) as well as Iso. dastychi (Bertolani et al. Citation2014a), Dor. macrodon, Dor. flavus, Fra. verrucosus requires further research. All these species were probably (a) wrongly identified or (b) belonged to other and probably not recognized new genera, as in case of Thu. augusti and/or Iso. dastychi (). Some nuclear sequences deposited in GenBank as Ere. alicatai could belong to an undefined species of the genus Grevenius or other undefined taxon. As recently indicated by Gąsiorek et al. (Citation2019b), the sequence HQ604951 of Ere. alicatai could be mislabeled. However, we showed that other sequences of Ere. alicatai, i.e. FJ435722 and FJ435767 could be misidentified and the sequence HQ604951 was clustered with newly sequenced Eremobiotus sp. We are not convinced which of these sequences have been correctly flagged and all of them seems to be questionable and need to be verify again. What is more, we also found that sequences designated as Thu. augusti were more similar to Pseudobiotus, Eremobiotus, Grevenius, Doryphoribius, Apodibius, Haplomacrobiotus and Hexapodibius than to other sequences identified as Thulinius Bertolani, Citation2003 (). In turn, sequences of Iso. dastychi were most similar to Ere. alicatai (but only to the one sequence of 18SrRNA (GenBank accession number: HQ604954)) and Eremobiotus sp. and it was also congruent with previous literature data (see Gąsiorek et al. Citation2019b). This is not surprising because the genus Isohypsibius still remains polyphyletic and it has been shown that Iso. dastychi represents a different evolutionary lineage within Isohypsibiidae (Gąsiorek et al. Citation2019b). In addition, we indicated that in the family Doryphoribiidae two species of the genus Doryphoribius, i.e. Dor. macrodon and Dor. flavus clustered in different subclades, so that they could represent genera, and their sequences need to be verified. Finally, Fra. verrucosus was placed within the clade Isohypsibius, which was also reported earlier by Gąsiorek et al. (Citation2019b).
Supplemental Material
Download MS Excel (39.8 KB)Supplemental Material
Download MS Excel (15 KB)Acknowledgements
Studies have been conducted in the framework of activities of BARg (Biodiversity and Astrobiology Research group).
Disclosure statement
No potential conflict of interest was reported by the authors.
Supplementary material
Supplemental data for this article can be accessed here.
References
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic local alignment search tool. Journal of Molecular Biology 215(3):403–410. DOI:10.1016/S0022-2836(05)80360-2.
- Beasley CW. 1995. The phylum Tardigrada. Abilene: USA. p. 1–1014. Third Edition by G. Ramazzotti and W. Maucci, English Translation. Published by the translator Clark Beasley.
- Beasley CW, Kaczmarek Ł, Michalczyk Ł. 2008. Doryphoribius mexicanus, a new species of Tardigrada (Eutardigrada: Hypsibiidae) from Mexico (North America). Proceedings of the Biological Society of Washington 121(1):34–40. DOI:10.2988/07-30.1.
- Bertolani R. 1982. 15. Tardigradi. Guide per il riconoscimento delle specie animali delle acque interne Italiane. Verona, Italy: Consiglio Nazionale Delle Ricerche. pp. 104.
- Bertolani R. 2003. Thulinius, new generic name substituting for Thulinia Bertolani, 1981 (Tardigrada, Eutardigrada). Zootaxa 314(1):1–4. DOI:10.11646/zootraxa.314.1.1.
- Bertolani R, Bartels PJ, Guidetti R, Cesari M, Nelson DR. 2014b. Aquatic tardigrades in the Great Smoky Mountains National Park, North Carolina and Tennessee, U.S.A., with the description of a new species of Thulinius (Tardigrada, Isohypsibiidae). Zootaxa 3764(5):524–536. DOI:10.11646/zootaxa.3764.5.2.
- Bertolani R, Guidetti R, Marchioro T, Altiero T, Rebecchi L, Cesari M. 2014a. Phylogeny of Eutardigrada: New molecular data and their morphological support lead to the identification of new evolutionary lineages. Molecular Phylogenetics and Evolution 76:110–126. DOI: 10.1016/j.ympev.2014.03.006.
- Binda MG. 1969. Nuovi dati su tardigradi di Sicilia con descrizione di due nuove specie. Bollettino delle Sedute della Accademia Gioenia di Scienze Naturali in Catania 9:623–633.
- Binda MG. 1980. Tardigradi di Lucania. Animalia 7:79–91.
- Binda MG, Pilato G. 1986. Ramazzottius, nuovo genere di Eutardigrado (Hypsibiidae). Animalia 13(1/3):159–166.
- Binda MG, Pilato G, Lisi O. 2005. Remarks on Macrobiotus furciger Murray, 1906 and description of three new species of the furciger group (Eutardigrada, Macrobiotidae). Zootaxa 1075(1):55–68. DOI:10.11646/zootaxa.1075.1.3.
- Casquet J, Thebaud C, Gillespie RG. 2012. Chelex without boiling, a rapid and easy technique to obtain stable amplifiable DNA from small amounts of ethanol-stored spiders. Molecular Ecology Resources 12(1):136–141. DOI:10.1111/j.1755-0998.2011.03073.x.
- Cesari M, McInnes S, Bertolani R, Rebecchi L, Guidetti R. 2016a. Genetic diversity and biogeography of the south polar water bear Acutuncus antarcticus (Eutardigrada: Hypsibiidae) – Evidence that it is a truly pan-Antarctic species. Invertebrate Systematics 30(6):635–649. DOI:10.1071/IS15045.
- Cesari M, Vecchi M, Palmer A, Bertolani R, Pilato G, Rebecchi L, Guidetti R. 2016b. What if the claws are reduced? Morphological and molecular phylogenetic relationships of the genus Haplomacrobiotus May, 1948 (Eutardigrada, Parachela). Zoological Journal of the Linnean Society 178(4):819–827. DOI:10.1111/zoj.12424.
- Ciobanu DA, Zawierucha K, Moglan I, Kaczmarek Ł. 2014. Milnesium berladnicorum sp. n. (Eutardigrada, Apochela, Milnesiidae), a new species of water bear from Romania. ZooKeys 429:1–11. DOI: 10.3897/zookeys.429.7755.
- Convey P, McInnes SJ. 2005. Exceptional tardigrade-dominated ecosystems in Ellsworth Land. Antarctica. Ecology 86(2):519–527. DOI:10.1890/04-0684.
- Dabert M, Dastych H, Dabert J. 2015. Molecular data support the dispersal ability of the glacier tardigrade Hypsibius klebelsbergi Mihelčič, 1959 across the environmental barrier (Tardigrada). Entomologische Mitteilungen aus dem Zoologischen Museum Hamburg 17:233–240.
- Dabert M, Dastych H, Hohberg K, Dabert J. 2014. Phylogenetic position of the enigmatic clawless eutardigrade genus Apodibius Dastych, 1983 (Tardigrada), based on 18S and 28S rRNA sequence data from its type species A. confusus. Molecular Phylogenetics and Evolution 70:70–75. DOI: 10.1016/j.ympev.2013.09.012.
- Darriba D, Taboada GL, Doallo R, Posada D. 2012. jModelTest 2: More models, new heuristics and parallel computing. Nature Methods 9:772. DOI: 10.1038/nmeth.2109.
- Dastych H. 1983. Apodibius confusus gen. n. sp. n. a new water-bear from Poland (Tardigrada). Bulletin of the Polish Academy of Sciences. Biological Sciences 31:41–46.
- Dastych H. 1984. The Tardigrada from Antarctica with description of several new species. Acta Zoologica Cracoviensia 27:377–436.
- Dastych H. 2018. Redescription and revalidation of the sub-Antarctic tardigrade Hypsibius murrayi (Richters, 1907) based on the rediscovered type material (Tardigrada, Panarthropoda). Entomologie heute 30:95–115.
- Degma P, Bertolani R, Guidetti. R 2020. Actual checklist of Tardigrada species. https://iris.unimore.it/retrieve/270444/Actual%20checklist%20of%20Tardigrada%2037th%20Edition%2008_07_20.pdf
- Degma P, Guidetti R. 2007. Notes to the current checklist of Tardigrada. Zootaxa 1579:41–53. DOI: 10.11646/zootaxa.1579.1.2.
- Doyère LMF. 1840. Memoire sur les Tardigrades. I. Annales Des Sciences Naturelles Paris 14:269–362.
- Folmer O, Black M, Hoeh W, Lutz R, Vrijenhoek R. 1994. DNA primers for amplification of mitochondrial cytochrome c oxidase subunit I from diverse metazoan invertebrates. Molecular Marine Biology and Biotechnology 3:294–299.
- Gąsiorek P, Michalczyk Ł. 2020. Revised Cornechiniscus (Heterotardigrada) and new phylogenetic analyses negate echiniscid subfamilies and tribes. Royal Society Open Science 7:200581. DOI: 10.1098/rsos.200581.
- Gąsiorek P, Morek W, Stec D, Blagden B, Michalczyk Ł. 2019a. Revisiting Calohypsibiidae and Microhypsibiidae: Fractonotus Pilato, 1998 and its phylogenetic position within Isohypsibiidae (Eutardigrada: Parachela). Zoosystema 41(6):71–89. DOI:10.5252/zoosystema2019v41a6.
- Gąsiorek P, Stec D, Morek W, Michalczyk Ł. 2018. An integrative redescription of Hypsibius dujardini (Doyère, 1840), the nominal taxon for Hypsibioidea (Tardigrada: Eutardigrada). Zootaxa 4415:45–75. DOI: 10.11646/zootaxa.4415.1.2.
- Gąsiorek P, Stec D, Morek W, Michalczyk Ł. 2019b. Deceptive conservatism of claws: Distinct phyletic lineages concealed within Isohypsibioidea (Eutardigrada) revealed by molecular and morphological evidence. Contributions to Zoology 88(1):78–132. DOI:10.1163/18759866-20191350.
- Gąsiorek P, Stec D, Morek W, Zawierucha K, Kaczmarek Ł, Lachowska-Cierlik D, Michalczyk Ł. 2016. An integrative revision of Mesocrista Pilato, 1987 (Tardigrada: Eutardigrada: Hypsibiidae). Journal of Natural History 50:2803–2828. DOI: 10.1080/00222933.2016.1234654.
- Gąsiorek P, Zawierucha K, Stec D, Michalczyk Ł. 2017. Integrative redescription of a common Arctic water bear Pilatobius recamieri (Richters, 1911). Polar Biology 40:2239–2252. DOI: 10.1007/s00300-017-2137-9.
- Goujon M, Mcwilliam H, Li W, Valentin F, Squizzato S, Paern J, Lopez R. 2010. A new bioinformatics analysis tools framework at EMBL–EBI. Nucleic Acids Resources 38:695–699. DOI: 10.1093/nar/gkq313.
- Guidetti R, Bertolani R. 2005. Tardigrade taxonomy: An updated check list of the taxa and a list of characters for their identification. Zootaxa 845:1–46. DOI: 10.11646/zootaxa.845.1.1.
- Guidetti R, McInnes SJ, Cesari M, Rebecchi L, Rota-Stabelli O. 2017. Evolutionary scenarios for the origin of an Antarctic tardigrade species based on molecular clock analyses and biogeographic data. Contributions to Zoology 86(2):97–110. DOI:10.1163/18759866-08602001.
- Guidetti R, Rebecchi L, Bertolani R, Jönsson KI, Kristensen RM, Cesari M. 2016. Morphological and molecular analyses on Richtersius (Eutardigrada) diversity reveal its new systematic position and lead to the establishment of a new genus and a new family within Macrobiotoidea. Zoological Journal of the Linnean Society 178:834–845. DOI: 10.1111/zoj.12428.
- Guidetti R, Rebecchi L, Cesari M, McInnes SJ. 2014. Mopsechiniscus franciscae, a new species of a rare genus of Tardigrada from continental Antarctica. Polar Biology 37(9):1221–1233. DOI:10.1007/s00300-014-1514-x.
- Guidetti R, Schill RO, Bertolani R, Dandekar T, Wolf M. 2009. New molecular data for tardigrade phylogeny, with the erection of Paramacrobiotus gen. n.. Journal of Zoological Systematics and Evolutionary Research 47:315–321. DOI: 10.1111/j.1439-0469.2009.00526.x.
- Guil N, Giribet G. 2012. A comprehensive molecular phylogeny of tardigrades – Adding genes and taxa to a poorly resolved phylum-level phylogeny. Cladistics 28:21–49. DOI: 10.1111/j.1096-0031.2011.00364.x.
- Guil N, Jørgensen A, Kristensen R. 2019. An upgraded comprehensive multilocus phylogeny of the Tardigrada tree of life. Zoologica Scripta 48:120–137. DOI: 10.1111/zsc.12321.
- Hall TA. 1999. BioEdit: A user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symposium Series 41:95–98. DOI: 10.14601/Phytopathol_Mediterr-14998u1.29.
- Iharos G. 1966. Neue Tardigraden-arten aus Ungarn. Acta Zoologica Academiae Scientiarum 12:111–122.
- Jørgensen A, Faurby S, Hansen JG, Mobjerg N, Kristensen RM. 2010. Molecular phylogeny of Arthrotardigrada (Tardigrada). Molecular Phylogenetics and Evolution 54(3):1006–1015. DOI:10.1016/j.ympev.2009.10.006.
- Jørgensen A, Kristensen RM. 2004. Molecular phylogeny of Tardigrada – Investigation of the monophyly of Heterotardigrada. Molecular Phylogenetics and Evolution 32:666–670. DOI: 10.1016/j.ympev.2004.04.017.
- Kaczmarek Ł, Bartylak T, Roszkowska M. 2020b. Two new genera of long clawed Isohypsibioidea Guil, Jørgensen & Kristensen, 2019. Zootaxa 4729(2):293–299. DOI:10.11646/zootaxa.4729.2.10.
- Kaczmarek Ł, Cytan J, Zawierucha K, Diduszko D, Michalczyk Ł. 2014. Tardigrades from Peru (South America), with descriptions of three new species of Parachela. Zootaxa 3790(2):357–379. DOI:10.11646/zootaxa.3790.2.5.
- Kaczmarek Ł, Grobys D, Kulpa A, Bartylak T, Kmita H, Kepel M, Kepel A, Roszkowska M. 2019. Two new species of the genus Milnesium Doyère, 1840 (Tardigrada, Apochela, Milnesiidae) from Madagascar. ZooKeys 884:1–22. DOI: 10.3897/zookeys.884.29469.
- Kaczmarek Ł, Michalczyk Ł. 2017. The Macrobiotus hufelandi group (Tardigrada) revisited. Zootaxa 4363(1):101–123. DOI:10.11646/zootaxa.4363.1.4.
- Kaczmarek Ł, Mioduchowska M, Kačarević U, Kubska K, Parnikoza I, Gołdyn B, Roszkowska M. 2020a. New records of Antarctic Tardigrada with comments on interpopulation variability of the Paramacrobiotus fairbanksi Schill, Förster, Dandekar and Wolf, 2010. Diversity 12:108. DOI: 10.3390/d12030108.
- Kaczmarek Ł, Parnikoza I, Gawlak M, Esefeld J, Peter H-U, Kozeretska I, Roszkowska M. 2018. Tardigrades from Larus dominicanus Lichtenstein, 1823 nests on the Argentine Islands (maritime Antarctic). Polar Biology 41(2):283–301. DOI:10.1007/s00300-017-2190-4.
- Kiehl E, Dastych H, D’Haese J, Greven H. 2007. The 18S rDNA sequences support polyphyly of Hypsibiidae (Eutardigrada). Journal of Limnology 66:21–25. DOI: 10.4081/jlimnol.2007.s1.21.
- Kihm JH, Kim S, McInnes SJ, Zawierucha K, Rho HS, Kang P, Park TYS. 2020. Integrative description of a new Dactylobiotus (Eutardigrada: Parachela) from Antarctica that reveals an intraspecific variation in tardigrade egg morphology. Scientific Reports 10:9122. DOI: 10.1038/s41598-020-65573-1.
- Kimble JM. 2004. Cryosols. Permafrost-affected soils. Berlin: Springer.
- Kumar S, Stecher G, Li M, Knyaz C, Tamura K. 2018. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Molecular Biology and Evolution 35(6):1547–1549. DOI:10.1093/molbev/msy096.
- Librado P, Rozas J. 2009. DnaSP v5: A software for comprehensive analysis of DNA polymorphism data. Bioinformatics 25:1451–1452. DOI: 10.1093/bioinformatics/btp187.
- Lindsay DC. 1971. Vegetation of the South Shetland Islands. British Antarctic Survey Bulletin 25:59–83.
- McInnes SJ. 2010. Echiniscus corrugicaudatus (Heterotardigrada; Echiniscidae) a new species from Ellsworth Land, Antarctica. Polar Biology 33:59–70. DOI: 10.1007/s00300-009-0684-4.
- Michalczyk Ł, Kaczmarek Ł. 2013. The Tardigrada Register: A comprehensive online data repository for tardigrade taxonomy. Journal of Limnology 72:175–181. DOI: 10.4081/jlimnol.2013.s1.e22.
- Mironov SV, Dabert J, Dabert M. 2012. A new feather mite species of the genus Proctophyllodes Robin, 1877 (Astigmata: Proctophyllodidae) from the long–tailed tit Aegithalos caudatus (Passeriformes: Aegithalidae): Morphological description with DNA barcode data. Zootaxa 3253:54–61. DOI: 10.11646/zootaxa.3253.1.2.
- Møbjerg N, Jørgensen A, Eibye-Jacobsen J, Halberg KA, Persson D, Kristensen RM. 2007. New records on cyclomorphosis in the marine eutardigrade Halobiotus crispae (Eutardigrada: Hypsibiidae). Journal of Limnology 66:132–140. DOI: 10.4081/jlimnol.2007.s1.132.
- Morek W, Ciosek J, Michalczyk Ł. 2020. Description of Milnesium pentapapillatum sp. nov., with an amendment of the diagnosis of the order Apochela and abolition of the class Apotardigrada (Tardigrada). Zoologischer Anzeiger 288:107–117. DOI: 10.1016/j.jcz.2020.07.002.
- Morek W, Gąsiorek P, Stec D, Blagden B, Michalczyk Ł. 2016a. Experimental taxonomy exposes ontogenetic variability and elucidates the taxonomic value of claw configuration in Milnesium Doyère, 1840 (Tardigrada: Eutardigrada: Apochela). Contributions to Zoology 85:173–200. DOI: 10.1163/18759866-08502003.
- Morek W, Stec D, Gąsiorek P, Schill RO, Kaczmarek Ł, Michalczyk Ł. 2016b. An experimental test of tardigrade preparation methods for light microscopy. Zoological Journal of the Linnean Society 178:785–793. DOI: 10.1111/zoj.12457.
- Morgan CI. 1976. Studies on the British tardigrade fauna. Some zoogeographical and ecological notes. Journal of Natural History 10:607–632. DOI: 10.1080/00222937600770491.
- Murray J. 1907. Scottish Tardigrada collected by the Lake Survey. Transactions of the Royal Society of Edinburgh 45:641–668. DOI: 10.1017/S0080456800011777.
- Nelson DR, Guidetti R, Rebecchi L. 2015. Chapter 17: Phylum Tardigrada. In: Thorp JH, Covich AP, editors. Ecology and general biology: vol. 1: freshwater invertebrates. 4th ed. London: Elsevier. pp. 347–380.
- Perry E, Miller WR, Kaczmarek Ł. 2019. Recommended abbreviations for the names of genera of the phylum Tardigrada. Zootaxa 4608:145–154. DOI: 10.11646/zootaxa.4608.1.8.
- Pilato G. 1974. Tardigradi delle acque dolci siciliane - Terza Nota. Animalia 1:235–244.
- Pilato G. 1981. Analisi di nuovi caratteri nello studio degli Eutardigradi. Animalia 8:51–57.
- Pilato G. 2013. The taxonomic value of the structures for the insertion of the stylet muscles in the Eutardigrada, and description of a new genus. Zootaxa 3721(4):365–378. DOI:10.11646/zootaxa.3721.4.4.
- Pilato G, Bertolani R, Binda MG. 1982. Studio degli Isohypsibius del gruppo elegans (Eutardigrada, Hypsibiidae) con descrizione di due nuove specie. Animalia 9:185–198.
- Pilato G, Binda MG. 2010. Definition of families, subfamilies, genera and subgenera of the Eutardigrada and keys to their identification. Zootaxa 2404:1–52. DOI: 10.5281/zenodo.194138.
- Pilato G, McInnes SJ, Lisi O. 2012. Hebesuncus mollispinus (Eutardigrada, Hypsibiidae), a new species from maritime Antarctica. Zootaxa 3446:60–68. DOI: 10.11646/zootaxa.3446.1.4.
- Pilato G, Sabella G, D’Urso V, Lisi O. 2017. Two new species of Eutardigrada from Victoria Land, Antarctica. Zootaxa 4317(3):541–558. DOI:10.11646/zootaxa.4317.3.6.
- Posada D, Buckley TR. 2004. Model selection and model averaging in phylogenetics: Advantages of the AIC and Bayesian approaches over likelihood ratio tests. Systematic Biology 53:793–808. DOI: 10.1080/10635150490522304.
- Pugh PJA, Convey P. 2008. Surviving out in the cold: Antarctic endemic invertebrates and their refugia. Journal of Biogeography 35:2176–2186. DOI: 10.1111/j.1365-2699.2008.01953.x.
- Ramazzotti G, Maucci W. 1983. Il Phylum Tardigrada. III edizione riveduta e aggiornata. Memorie dell’Istituto Italiano di Idrobiologia 41:1–1012.
- Rambaut A, Suchard MA, Xie D, Drummond AJ. 2014. Tracer v1.6. Available from http://beast.bio.ed.ac.uk/Tracer
- Rice P, Longden I, Bleasby A. 2000. EMBOSS: The European molecular biology open software suite. Trends in Genetics 16:276–277. DOI: 10.1016/s0168-9525(00)02024-2.
- Richters F. 1900. Beiträge zur Kenntnis der fauna der Umgebung von Frankfurt a. Frankfurt a. M: M. Ber. Senkenberg. Naturf. Ges. 21–44 pp.
- Richters F. 1926. Tardigrada. In: Kükenthal W, Krumbach T, editors. Handbuch der Zoologie. Berlin: Walter de Gruyter & Co. Vol. 3. pp. 58–61.
- Riffenburh B. 2007. Encyclopedia of the Antarctic. New York: Taylor Francis Group.
- Ronquist F, Huelsenbeck JP. 2003. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19:1572–1574. DOI: 10.1093/bioinformatics/btg180.
- Sands CJ, Mcinnes SJ, Marley NJ, Goodall–Copestake WP, Convey P, Linse K. 2008. Phylum Tardigrada: An “individual” approach. Cladistics 24:861–871. DOI: 10.1111/j.1096-0031.2008.00219.x.
- Schill RO, Steinbrück G. 2007. Identification and differentiation of Heterotardigrada and Eutardigrada species by riboprinting. Journal of Zoological Systematics and Evolutionary Research 45:184–190. DOI: 10.1111/j.1439-0469.2007.00409.x.
- Schultze CAS. 1834. Macrobiotus Hufelandii animal e crustaceorum classe novum, reviviscendi post diuturnam asphixiam et aridiatem potens, etc. 8, 1 tab. C. Berlin: Curths. p. 6. I Table.
- Schuster RO, Nelson DR, Grigarick AA, Christenberry D. 1980. Systematic criteria of Eutardigrada. Transactions of the American Microscopical Society 99:284–303. DOI: 10.2307/3226004.
- Smith RIL, Corner RWM. 1973. Vegetation of the Arthur Harbour-Argentine Islands region of the Antarctic Peninsula. British Antarctic Survey Bulletin 33(34):89–122.
- Stec D, Morek W, Gąsiorek P, Michalczyk Ł. 2018. Unmasking hidden species diversity within the Ramazzottius oberhaeuseri complex, with an integrative redescription of the nominal species for the family Ramazzottiidae (Tardigrada: Eutardigrada: Parachela). Systematics and Biodiversity 16:357–376. DOI: 10.1080/14772000.2018.1424267.
- Stec D, Smolak R, Kaczmarek Ł, Michalczyk Ł. 2015. An integrative description of Macrobiotus paulinae sp. nov. (Tardigrada: Eutardigrada: Macrobiotidae: hufelandi group) from Kenya. Zootaxa 4052(5):501–526. DOI:10.11646/zootaxa.4052.5.1.
- Stec D, Zawierucha K, Michalczyk Ł. 2017. An integrative description of Ramazzottius subanomalus (Biserov, 1985) (Tardigrada) from Poland. Zootaxa 4300:403–420. DOI: 10.11646/zootaxa.4300.3.4.
- Thulin G. 1928. Über die Phylogenie und das System der Tardigraden. Hereditas Lund 11:207–266.
- Tsujimoto M, Kagoshima H, Kanda H, Watanabe K, Imura S. 2020. Reproductive performance of the Antarctic tardigrades, Acutuncus antarcticus (Eutardigrada: Hypsibiidae), revived after being frozen for over 30 years and of their offspring. Zoological Journal of the Linnean Society 188(3):839–847. DOI:10.1093/zoolinnean/zlz137.
- Tsujimoto M, McInnes SJ, Convey P, Imura S. 2014. Preliminary description of tardigrade species diversity and distribution pattern around coastal Syowa Station and inland Sør Rondane Mountains, Dronning Maud Land, East Antarctica. Polar Biology 37(9):1361–1367. DOI:10.1007/s00300-014-1516-8.
- Tumanov DV. 2003. Four new Isohypsibius species from Russian fresh waters (Tardigrada, Hypsibiidae). Bulletin De l’Institut Royal Des Sciences Naturelles De Belgique Biologie 73:183–189.
- Vecchi M, Cesari M, Bertolani R, Jönsson KI, Rebecchi L, Guidetti R. 2016. Integrative systematic studies on tardigrades from Antarctica identify new genera and new species within Macrobiotoidea and Echiniscoidea. Invertebrate Systematics 30(4):303–322. DOI:10.1071/IS15033.
- Velasco-Castrillón A, Gibson JAE, Stevens MI. 2014. A review of current Antarctic limno-terrestrial microfauna. Polar Biology 37(10):1517–1531. DOI:10.1007/s00300-014-1544-4.
- Vicente F, Bertolani R. 2013. Considerations on the taxonomy of the Phylum Tardigrada. Zootaxa 3626(2):245–248. DOI:10.11646/zootaxa.3626.2.2.
- Zawierucha Z, Stec D, Lachowska-Cierlik D, Takeuchi N, Li Z, Michalczyk Ł. 2018. High mitochondrial diversity in a new water bear species (Tardigrada: Eutardigrada) from mountain glaciers in central Asia, with the erection of a new genus Cryoconicus. Annales Zoologici 68(1):179–201. DOI:10.3161/00034541ANZ2018.68.1.007.

