Abstract
The only Palearctic representative of the leech genus Placobdella Blanchard, 1893 is P. costata, an ectoparasite of freshwater turtles. To date, no conclusive evidence about the possible presence of coevolutionary patterns between this leech and its turtle hosts is available due to the paucity of DNA sequence data available for P. costata; moreover, comparative host data is also mostly lacking, making any inferences more difficult. The discovery of new populations of the species in northern Italy and Sicily allowed us to generate novel mitochondrial DNA sequences and to compare the topology of the resulting phylogenetic trees with the phylogeny of the turtle hosts occurring in the study area, i.e., Emys orbicularis and E. trinacris. The branching pattern of the phylogenetic tree for P. costata is not congruent with that of its turtle hosts, thus suggesting the lack of coevolutionary or cospeciation phenomena between these taxa. The lack of a coevolutionary pattern might be ascribed to the different dispersal ability of Placobdella costata and Emys spp. and to the host generality of the leech, as confirmed by the occurrence of P. costata on aquatic turtles belonging to the genus Mauremys in areas where Emys spp. are rare or absent.
A single leech cytochrome c oxidase subunit 1 haplotype was found in each study region, and the overall nucleotide diversity was very low throughout the investigated distribution. This apparent lack of a clear phylogeographical pattern was unexpected in the P. costata populations occurring in the circum-Mediterranean areas, where the occurrence of high haplotype and nucleotide diversity is customary for most terrestrial and freshwater species. Based on the available data, we suggest a recent, post-glacial origin of the studied P. costata populations.
Introduction
Cospeciation and coevolution are assumed to often occur in host-parasite systems although the actual frequency of real cospeciation patterns has recently been shrouded in doubt (De Vienne et al. Citation2013). In ideal cases, the cospeciation should follow the “Fahrenholz rule”, i.e., the phylogeny of permanent and specialised parasites should follow that of the host taxa (Fahrenholz Citation1913). However, exceptions are known, and these are mostly ascribed to host switching, intra-host speciation or parasite inertia (Paterson & Banks Citation2001; Legendre et al. Citation2002). Based on their shared evolutionary history characterised by the colonisation of the Palearctic from the Nearctic region (Siddall et al. Citation2005), and on the allegedly close trophic relationships that occur between them (Minelli Citation1979; Nesemann & Neubert Citation1999), a cospeciation pattern could be expected between the leech Placobdella costata and its emydine turtle hosts Emys spp. Instead, the exploratory data produced by Marrone et al. (Citation2016) rather suggested the existence of independent evolutionary patterns in these taxa or a possible human-mediated introduction of Placobdella costata in Sicily from the southern Italian peninsula (Naselli-Flores & Marrone Citation2019). However, the inclusion of P. costata samples originating from a single locality in each investigated region did not provide definitive evidence, and further in-depth surveys were strongly advocated by the authors.
The recent findings of new Italian P. costata populations in Sicily and Piedmont (Evangelista & Seglie Citation2016; F. Marrone, unpubl. data) and the deposition of some European and Asian P. costata sequences in the publicly accessible GenBank database provided an opportunity to better investigate the molecular diversity pattern of this rare leech species in Italy, and to compare it with the phylogeny of its sympatric hosts, i.e., freshwater turtles belonging to Emys trinacris, E. orbicularis galloitalica and E. orbicularis hellenica (Vamberger et al. Citation2015).
Materials and methods
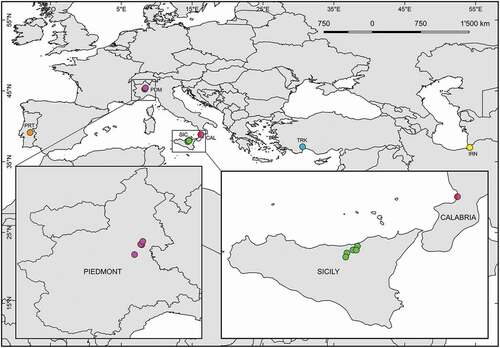
Placobdella costata individuals were collected from their turtle hosts in novel occurrence sites located in Piedmont and Sicily (Italy) () preserved in situ in 90% ethanol, and identified according to Minelli (Citation1977, Citation1979) and Nesemann and Neubert (Citation1999). Details about the sampled localities are reported in . Eight P. costata specimens from Sicily and Piedmont were deposited at the Museo di Storia Naturale, Sezione di Zoologia “La Specola”, Università di Firenze (S.M.A.), Italy under the collection numbers MZUF AN/6922/6154 - AN/6927/6159; further, specimens are stored in the authors’ collection at the University of Palermo, Italy, and are available for loan on request. When enough specimens were available, total DNA was extracted from two P. costata individuals from each site, and a fragment of the mitochondrial gene cytochrome c oxidase subunit 1 (COI) was amplified following the procedures described in Marrone et al. (Citation2016), sequenced with an ABI 3130xL sequencer (Applied Biosystems) by Macrogen SPAIN, and uploaded to the public database GenBank under accession numbers MW935927-MW935939.
Table I. Origin and GenBank Accession Numbers (A.N.) of the analysed Placobdella costata specimens. Geographical coordinates are expressed in terms of decimal degrees (Map Datum: WGS84)
Figure 1. Geographic location of the sampled sites. See for their coordinates and information on the collected Placobdella costata specimens and their hosts. Codes refer to those listed in . The geographic coordinates of the Portuguese and Iranian sites are unknown; accordingly, orange and yellow dots show their approximate locations
In addition, all the P. costata COI sequences available on GenBank on 15/04/2021 and a sequence of P. ornata (Verrill, 1872) to be used as outgroups were downloaded from GenBank and included in the analyses (“Placobdella dataset”; see and ) for their Accession Numbers, AN). All sequences were aligned using the software MEGAX (Kumar et al. Citation2018) with the ClustalW method (Thompson et al. Citation1994). Pairwise haplotype uncorrected p-distances were calculated with MEGAX (Kumar et al. Citation2018).
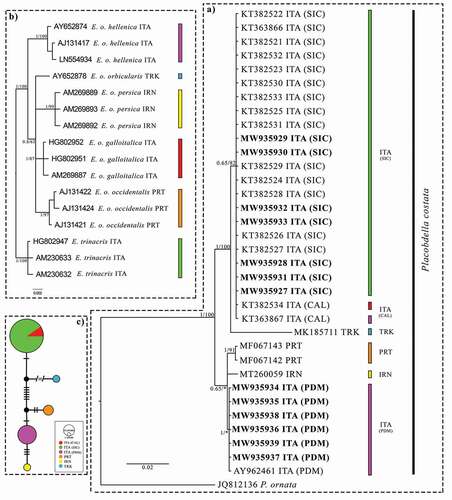
Figure 2. (a) Bayesian phylogram (95% majority rule consensus tree) of Placobdella costata based on a 540 bp fragment of the mtDNA COI. A sequence of P. ornata (A.N. JQ812136) was used as an outgroup to root the tree. See for the geographic origin of each sequence. Novel sequences are reported in bold. Node statistical support is reported as nodal posterior probabilities (Bayesian Inference of phylogeny, BI)/bootstrap values (Maximum Likelihood, ML). Asterisks indicate a bootstrap support value lower than 50. The colours of the pie chart refer to those shown in Figure 2(b) and refer to the hosts (Emys spp.) of the parasite (P. costata) in each analysed area. (b) Bayesian phylogram (95% majority rule consensus tree) of Emys spp. based on a 1031 bp fragment of the mtDNA Cytb. Node statistical support is reported as nodal posterior probabilities (Bayesian Inference of phylogeny, BI)/bootstrap values (Maximum Likelihood, ML). (c) Median-joining haplotype network based on the mtDNA COI of Placobdella costata. Dashes indicate substitutions steps. Each circle represents a haplotype, and its size is proportional to its frequency. All the analysed specimens are reported using the codes listed in
The phylogenetic relationships among the studied populations were investigated using Bayesian inference (BI) and maximum likelihood (ML) methods as implemented in the software packages MrBayes v. 3.2.7 (Ronquist et al. Citation2012) and PhyML v. 3 (Guindon & Gascuel Citation2003). In accordance with the best evolutionary model selected by PartitionFinder v. 1.0.1 (Lanfear et al. Citation2012), both BI and ML analyses were performed under a General Time Reversible model of sequence evolution with a proportion of invariable sites (GTR+I). As a measure of branch support, bootstrap values were calculated with 1000 replicates in the ML trees. For the BI analysis, two independent Markov Chain Monte Carlo (MCMC) analyses were run with 1.000.000 generations (temp.: 0.2; default priors). The convergence of the runs was assessed with Tracer v1.6 (Rambaut et al. Citation2014), obtaining effective sample size (ESS) values above 200 for all parameters.
In order to compare the branching pattern of Placobdella costata trees with that of its Italian hosts, selected Emys spp. cytochrome b (cytb) sequences were downloaded from GenBank. Since comparative GenBank P. costata sequences from Portugal, Iran and Turkey were included in the “Placobdella dataset”, cytb sequences of Emys orbicularis occidentalis, E. orbicularis persica and Emys orbicularis orbicularis from these countries were included in the “Emys dataset” as well (see ) for their Accession Numbers). The BI and ML analyses of the “Emys dataset” were performed under a Hasegawa–Kishino–Yano model of evolution with a proportion of invariable sites (HKY + I) implementing the settings reported above for the “Placobdella dataset”.
A haplotype network including all Placobdella costata sequences were constructed based on the “Placobdella dataset” using the software PopART v. 1.7 (http://popart.otago.ac.nz) implementing the median-joining network algorithm as suggested by Bandelt et al. (Citation1999).
Results
New P. costata populations were found in four sites in Sicily and three sites in Piedmont. The number of investigated leeches per site and their turtle hosts are reported in .
Overall, a 577-bp long COI fragment was successfully amplified and sequenced from 13 P. costata individuals originating from seven novel sites; novel sequences were aligned with the further 21 COI P. costata sequences and a single P. ornata sequence downloaded from GenBank (). After having trimmed the sequence tails which are not present in all the sequences, a 540-bp long aligned COI fragment was obtained for the “Placobdella dataset”. All the sequenced P. costata individuals from Sicily presented the same haplotype already observed by Marrone et al. (Citation2016) in Sicily and southern Calabria, whereas all the new P. costata individuals from Piedmont showed the same COI haplotype already observed in a specimen from Piedmont by Siddall et al. (Citation2005), thus consistently highlighting a noteworthy homogeneity of the local populations of the species.
The phylogenetic tree rooted at Placobdella ornata ()) shows the presence of two well-supported clades, one including the single available Turkish sequence and the Italian sequences from Sicily and Calabria (southern Italy), the other including the sequences from Piedmont (northern Italy), Portugal, and Iran. The first clade is further divided into two subclades, including the sequences from P. costata individuals from southern Italy and from Turkey, respectively, the second clade is characterised by the presence of three haplotypes with unresolved relationships, each one related to sequences with different geographical origins (i.e., northern Italy, Portugal, and Iran). Oddly enough, the mtDNA diversity pattern observed in the “Placobdella dataset” does not show a large-scale geographical pattern and suggests close relationships between distantly located populations (e.g., among P. costata populations occurring in Portugal, northern Italy, and Iran) and vice-versa, a significant phylogenetic distance between geographically closer populations (i.e., among the northern vs. southern Italian study sites). The uncorrected pairwise distance occurring among the observed haplotypes is reported in .
Table II. Pairwise uncorrected p-distance between the cytochrome oxidase subunit 1 (COI) haplotypes observed in Placobdella costata. Codes refer to those listed in
A phylogenetic tree based on selected Emys spp. cytb sequences ()) is in general agreement with the phylogenetic relationships known to occur between Emys species and subspecies (e.g., Stuckas et al. Citation2014).
With regard to the geographic localities from which the specimens were collected, the branching patterns of the phylogenetic trees based on the “Placobdella dataset” and on the “Emys dataset” are incongruent since two distantly related P. costata lineages are found to occur on Emys orbicularis hellenica and, conversely, a single P. costata haplotype is shared by three different turtle hosts, i.e., E. trinacris, E. orbicularis galloitalica, and E. orbicularis hellenica. Such a result is further stressed by the haplotype network ()), which shows that a single COI P. costata haplotype is shared by different turtle hosts and, conversely, different and distantly related P. costata haplotypes can be observed in Emys orbicularis hellenica.
Discussion
The analysis of the “Placobdella dataset” showed the existence of distinct P. costata COI haplotypes and clades occurring in different areas although their phylogenetic relationships do not seem to follow any clear geographical pattern; the close relationship between the haplotype found in southern Italy and that found in Turkey or between the haplotype found in northern Italy and those found in Portugal and Iran, do not have any straightforward biogeographical explanation ()). Moreover, based on the available evidence, the phylogenetic relationships among the Italian P. costata populations are completely independent from those observed in their host taxa () since the host differentiation is not mirrored by a parallel diversification of the parasite. This pattern is here ascribed to the different dispersal ability of the two studied taxa, and to the non-exclusive link between the hosts and the parasite; conversely, the alternative hypothesis of a recent human-mediated introduction of P. costata in Sicily proposed by Marrone et al. (Citation2016) and Arizza et al. (Citation2016) is made unparsimonious by the available data since the incongruence between Emys spp. and P. costata phylogenies is apparent also for other geographical areas.
The dispersal ability of freshwater turtles is relatively limited and, with few exceptions (e.g., Vamberger et al. Citation2014), it is deeply influenced by sea straits and mountain ranges, e.g., in a recent study carried out in Sicily, Vecchioni et al. (Citation2020) showed that different E. trinacris independent management units can be singled out even at the scale of few tens or hundreds of kilometres, in accordance with the known philopatry of aquatic turtles. Conversely, Placobdella spp. can be passively dispersed over long ranges by biological vectors (Davies et al. Citation1982; Vamberger & Trontelj Citation2007); thus, presenting a molecular diversity pattern which is expected to mirror that of its avian or mammal dispersal vectors rather than those of its primary hosts. Moreover, a growing evidence shows that the alleged strict link between P. costata and aquatic turtles belonging to the genus Emys was overestimated, since the species routinely occurs on Mauremys spp. (e.g., Romero et al. Citation2014; Bashirichelkasari & Yadollahvandmiandoab Citation2017; Laghzaoui et al. Citation2020) where Emys hosts are rare or absent. Cospeciation requires tight physiological interactions between hosts and parasites, and the acquisition of new hosts, or the parasite dispersal mediated by alternative hosts are known to prevent cospeciation or coevolutionary phenomena from occurring (Ricklefs et al. Citation2004). In light of the obtained results, the existence of cospeciation or coevolutionary patterns between the Italian P. costata populations and their turtle hosts can likely be ruled out, and the possible non-native status of P. costata in Sicily is not supported.
A strikingly low mtDNA diversity was observed for the leech species at both Italian and West Palearctic scales, which contrasts with the high genetic diversity expected to occur in the circum-Mediterranean countries. In fact, southern European peninsulas acted as refugia during the Pleistocene glacial cycles, allowing the persistence of ancient local haplotypes and ultimately bringing to a high local haplotype diversity belonging to endemic haplogroups in each refugial area (e.g., Hewitt Citation2004; Arias et al. Citation2021). Conversely, the observed pattern shows the existence of a few widely spread and closely related haplotypes, with a negligible diversity occurring at a local scale. This pattern suggests a post-glacial (re)colonization of the study areas by P. costata originating from an unknown source area as already observed in the Italian peninsula and Sicily in the freshwater crab Potamon fluviatile (Herbst, 1785) (Vecchioni et al. Citation2017; Marrone et al. Citation2020). The causes underlying the current molecular diversity patterns of P. costata are to date unclear, and a dedicated study to explore the phylogeography of the species throughout its distribution range is desirable.
Author contribution
FM, VA and MA conceived the idea and planned the methods. LV, ML, DS, RC, GB and FM collected and analysed the data. LV and FM wrote the manuscript. All authors provided a critical revision to the final manuscript.
Acknowledgements
The authors wish to thank R. Scardino (Italy) for the help she provided in the frame of the field work in Sicily. S. Cianfanelli (University of Florence, Italy) kindly provided the catalogue numbers of the specimens deposited in the collection of the Museo di Storia Naturale, Sezione di Zoologia “La Specola” (Florence, Italy). Two anonymous reviewers are acknowledged for their constructive criticisms.
Disclosure statement
No potential conflict of interest was reported by the authors.
References
- Arias A, Surugiu V, Carballeira R, Popa OP, Popa LO, Utevsky S. 2021. Unravelling the extent of diversity within the Iberian medicinal leeches (Hirudinea: Hirudo) using molecules and morphology. Biology 10(4):315. DOI: 10.3390/biology10040315.
- Arizza V, Sacco F, Russo D, Scardino R, Arculeo M, Vamberger M, Marrone F. 2016. The good, the bad and the ugly: Emys trinacris, Placobdella costata and Haemogregarina stepanowi in Sicily (Testudines, Annelida and Apicomplexa). Folia Parasitologica 63:029. DOI: 10.14411/fp.2016.029.
- Bandelt HJ, Forster P, Röhl A. 1999. Median-joining networks for inferring intraspecific phylogenies. Molecular Biology and Evolution 16(1):37–48. DOI: 10.1093/oxfordjournals.molbev.a026036.
- Bashirichelkasari N, Yadollahvandmiandoab R. 2017. Placobdella costata an ectoparasite for Mauremys caspica in north of Iran. Journal of Aquaculture Research and Development 8:506. DOI: 10.4172/2155-9546.1000506.
- Davies RW, Linton LR, Wrona FJ. 1982. Passive dispersal of four species of freshwater Leeches (Hirudinoidea) by Ducks. Freshwater Invertebrate Biology 1:40–44. DOI: 10.2307/1467140.
- De Carle D, Oceguera-Figueroa A, Tessler M, Siddal ME, Kvist S. 2017. Phylogenetic analysis of Placobdella (Hirudinea: Rhynchobdellida: Glossiphoniidae) with consideration of COI variation. Molecular Phylogenetics and Evolution 114:234–248. DOI: 10.1016/j.ympev.2017.06.017.
- De Vienne DM, Refrégier G, López‐Villavicencio M, Tellier A, Hood ME, Giraud T. 2013. Cospeciation vs host‐shift speciation: Methods for testing, evidence from natural associations and relation to coevolution. New Phytologist 198:347–385. DOI: 10.1111/nph.12150.
- Evangelista M, Seglie D. 2016. Primo ritrovamento in Piemonte di Placobdella costata (Fr. Müller, 1846) (Annelida, Hirudinida, Glossiphoniidae). Rivista piemontese di storia naturale 37:49–57.
- Fahrenholz H. 1913. Ectoparasiten und Abstammungslehre. Zoologischer Anzeiger 41:371–374.
- Guindon S, Gascuel O. 2003. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Systematic Biology 52:696–704. DOI: 10.1080/10635150390235520.
- Hewitt G. 2004. Genetic consequences of climatic oscillations in the quaternary. Philosophical Transactions of the Royal Society B 359:183–195. DOI: 10.1098/rstb.2003.1388.
- Kumar S, Stecher G, Li M, Knyaz C, Tamura K. 2018. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Molecular Biology and Evolution 35(6):1547–1549. DOI: 10.1093/molbev/msy096.
- Laghzaoui EM, Abbad A, El Mouden E. 2020. Host-parasite association of Placobdella costata (Glossiphoniidae: Hirudinea) and Mauremys leprosa (Geoemydidae: Testudinoidea) in aquatic ecosystems of Morocco. Parasitology Research 119:3459–3467. DOI: 10.1007/s00436-020-06809-x.
- Lanfear R, Calcott B, Ho SYW, Guidon S. 2012. PartitionFinder: Combined selection of partitioning schemes and substitution models for phylogenetic analyses. Molecular Biology and Evolution 29:1695–1701. DOI: 10.1093/molbev/mss020.
- Legendre P, Desdevises Y, Bazin E. 2002. A statistical test for host-parasite coevolution. Systematic Biology 51:217–234. DOI: 10.1080/10635150252899734.
- Marrone F, Sacco F, Kehlmaier C, Arizza V, Arculeo M. 2016. Some like it cold: The glossiphoniid parasites of the Sicilian endemic pond turtle Emys trinacris (Testudines, Emydidae), an example of ‘parasite inertia’? Journal of Zoological Systematics and Evolutionary Research 54:60–66. DOI: 10.1111/jzs.12117.
- Marrone F, Vecchioni L, Deidun A, Mabrouki Y, Arab A, Arculeo M. 2020. DNA taxonomy of the potamid freshwater crabs from Northern Africa (Decapoda, Potamidae). Zoologica Scripta 49:473–487. DOI: 10.1111/zsc.12415.
- Minelli A 1977. Irudinei (Hirudinea). Guide per il Riconoscimento delle Specie Animali delle Acque Interne Italiane. CNR AQ/1/2. pp. 42.
- Minelli A. 1979. Fauna d’Italia 15: Hirudinea. Bologna: Calderini. pp. 152.
- Naselli-Flores L, Marrone F. 2019. Different invasibility of permanent and temporary waterbodies in a semiarid Mediterranean Island. Inland Waters 9:411–421. DOI: 10.1080/20442041.2019.1653110.
- Nesemann H, Neubert E. 1999. Anellida, Clitellata: Brachiobdellida, Acanthobdellea, Hirudinea. Berlin: Spektrum Akademisher Verlag Heideberg.
- Paterson AM, Banks J. 2001. Analytical approaches to measuring cospeciation of host and parasites: Through a glass, darkly. International Journal for Parasitology 31:1012–1022. DOI: 10.1016/s0020-7519(01)00199-0.
- Rambaut A, Suchard MA, Xie D, Drummond AJ. 2014. Tracer v.1.6. Available: http://beast.bio.ed.ac.uk/Tracer
- Ricklefs RE, Fallon SM, Bermingham E. 2004. Evolutionary relationships, cospeciation, and host switching in Avian Malaria Parasites. Systematic Biology 53(1):111–119. DOI: 10.1080/10635150490264987.
- Romero D, Duarte J, Narváez-Ledesma L, Farfán M, Real R. 2014. Presence of the leech Placobdella costata in the south of the Iberian Peninsula. Acta Parasitologica 59(2):259–262. DOI: 10.2478/s11686-014-0232-4.
- Ronquist F, Teslenko M, Van Der Mark P, Ayres DL, Darling A, Höhna S, Huelsenbeck JP. 2012. MrBayes 3.2: Efficient Bayesian phylogenetic inference and model choice across a large model space. Systematic Biology 61(3):539–542. DOI: 10.1093/sysbio/sys029.
- Siddall ME, Budinoff RB, Borda E. 2005. Phylogenetic evaluation of systematics and biogeography of the leech family Glossiphoniidae. Invertebrate Systematics 19:105–112. DOI: 10.1071/IS04034.
- Stuckas H, Velo-Antón G, Fahd S, Kalboussi M, Rouag R, Arculeo M, Marrone F, Sacco F, Vamberger M, Fritz U. 2014. Where are you from, stranger? The enigmatic biogeography of North African pond turtles (Emys orbicularis). Organisms Diversity & Evolution 14:295–306. DOI: 10.1007/s13127-014-0168-4.
- Thompson JD, Higgins DG, Gibson TJ. 1994. CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Research 22(22):4673–4680. DOI: 10.1093/nar/22.22.4673.
- Vamberger M, Stuckas H, Ayaz D, Lymberakis P, Široký P, Fritz U. 2014. Massive transoceanic gene flow in a freshwater turtle (Testudines: Geoemydidae: Mauremys rivulata). Zoologica Scripta 43:313–322. DOI: 10.1111/zsc.12055.
- Vamberger M, Stuckas H, Sacco F, D’Angelo S, Arculeo M, Cheylan M, Corti C, Lo Valvo M, Marrone F, Wink M, Fritz U. 2015. Differences in gene flow in a twofold secondary contact zone of pond turtles in southern Italy (Testudines: Emydidae: Emys orbicularis galloitalica, E. o. hellenica, E. trinacris). Zoologica Scripta 44:233–249. DOI: 10.1111/zsc.12102.
- Vamberger M, Trontelj P. 2007. Placobdella costata (Fr. Müeller, 1846) (Hirudinea: Glossiphoniidae), a leech species new for Slovenia. Natura Sloveniae 9:37–42.
- Vecchioni L, Deidun A, Sciberras J, Sciberras A, Marrone F, Arculeo M. 2017. The late Pleistocene origin of the Italian and Maltese populations of Potamon fluviatile (Malacostraca: Decapoda): Insights from an expanded sampling of molecular data. The European Zoological Journal 2017:575–582. DOI: 10.1080/24750263.2017.1405084.
- Vecchioni L, Marrone F, Arculeo M, Fritz U, Vamberger M. 2020. Stand out from the crowd: Small-scale genetic structuring in the endemic Sicilian pond turtle. Diversity 12(9):343. DOI: 10.3390/d12090343.
