Abstract
Niphargus comprises hundreds of narrowly-endemic West Palaearctic subterranean taxa. However, a few exceptional species inhabit surface waters and have remarkably large ranges. Herein, based on morphological and molecular analyses, we provide important new records for two of these species. N. potamophilus, previously known from the eastern Azov Sea lowlands, is reported for the first time from Ukraine and Bulgaria from localities adjacent to the Black Sea. These findings expand its range westward by more than 1000 km along the coastline. From Bulgaria, we also report for the first time N. hrabei, a species previously known to occur along the middle and lower Danube lowlands and in isolated populations at the foothills of the Northern Caucasus. Our new record thus extends its range southwards by more than 150 km. Both species contained unique haplotypes at all of the sampled localities. These were, nevertheless, not very divergent from more distant populations, emphasizing their good dispersal ability. Ecologically, the sampling localities were generally characterized by stagnant to low running water, dense vegetation, and muddy substrate. Overall, our results bring important insights, shedding more light on the biogeography and ecology of Niphargus.
Keywords:
Introduction
The genus Niphargus Schiödte, 1849 is characterized by high species diversity (>400) which is distributed across Europe and West Asia (Horton et al. Citation2022). Although most species have small ranges (<200 km), some taxa show a wide distribution (Copilaș-Ciocianu et al. Citation2017). However, many of the large ranged species turned out to be complexes of cryptic species with smaller ranges (Lefébure et al. Citation2006, Citation2007; Trontelj et al. Citation2009). Most Niphargus live in hypogean (subterranean) waters (Fišer Citation2012), but few inhabit the ecotone zone between the hypogean and epigean (surface) realms (Straškraba Citation1972; Copilaş-Ciocianu et al. Citation2014; Copilaș-Ciocianu et al. Citation2017).
Remarkably, a few species of Niphargus are known almost exclusively from epigean habitats, and they also have the widest known ranges of any other niphargid, despite being blind and depigmented (Copilaș-Ciocianu et al. Citation2017). The best known are Niphargus valachicus Dobreanu & Manolache, 1933 and N. hrabei S. Karaman, 1932, both inhabiting the middle and lower Danube lowlands as well as being occasionally reported around the Black Sea lowlands at the foothills of the Caucasus or the Anatolian Peninsula (Cărăușu et al. Citation1955; Nesemann et al. Citation1995; Copilaş-Ciocianu et al. Citation2014; Copilaș-Ciocianu et al. Citation2017; Palatov & Marin Citation2021). Although both are similar from a morphological, ecological and biogeographic perspective, they are not closely related (Copilaş-Ciocianu et al. Citation2018; Palatov & Marin Citation2021).
Niphargus potamophilus (Birstein Citation1954) is another lowland species that inhabits similar habitats and has a similar morphology to N. hrabei and N. valachicus, being closely related to the latter (Birstein Citation1954; Palatov & Marin Citation2021). These species have trapezoidal gnathopods, acutely produced infero-posterior corners of epimeres 2 and 3, additional spines on pereopod 3–4 dactily, non-sexually dimorphic 3rd uropod and a spoon-shaped appendix on the distal end of the uropod 1 peduncle. Niphargus potamophilus was originally described based on the specimens collected from the Don River delta, from a floodplain reservoir nearby Rostov-on-Don City, and from fish farms in Atkhyr, Kuban River delta, ca. 300 km southwest from the Don delta (Birstein Citation1954). The recognized range of this species is much smaller than that of N. hrabei and N. valachicus, for now being reported along the eastern Azov Sea shore (Palatov & Marin Citation2021), but it was suggested that its distribution might be much wider, covering southern Ukrainian rivers (Birstein Citation1954).
Niphargus hrabei, N. potamophilus and N. valachicus are epigean species, rarely, if at all, being reported from groundwater (Karaman Citation1950; Dedju Citation1967; Sket Citation1981; Meijering et al. Citation1995; Nesemann et al. Citation1995; Copilaş-Ciocianu et al. Citation2014; Copilaș-Ciocianu et al. Citation2017; Palatov & Marin Citation2021). Only N. hrabei is known to have a handful of stable subterranean populations (Dudich Citation1941; Meijering et al. Citation1995; Pérez-Moreno et al. Citation2017). All three species inhabit the stagnant/slow-flowing, densely vegetated, eutrophic waters with a muddy substrate of lowland springs, streams, canals, rivers, ponds, lakes, and temporary water bodies (Nesemann et al. Citation1995; Copilaş-Ciocianu et al. Citation2014; Copilaș-Ciocianu et al. Citation2017; Palatov & Marin Citation2021).
In the present study, we report N. potamophilus and N. hrabei from new territories and river basins in Bulgaria and Ukraine. Our combined molecular and morphological approach confirmed that these species are more widespread than previously known.
Material and methods
Sampling
Specimens were collected with a hand net during sampling campaigns in Ukraine in 2018 and in Bulgaria in 2014–2018 (for details see ). Animals were fixed in 96% ethanol. The material is stored in the collection of invertebrates at the Department of Invertebrate Zoology and Hydrobiology (University of Lodz, Poland) and Nature Research Center (Lithuania).
Table I. New localities and GenBank accession numbers of specimens were used for the current study.
DNA extraction, amplification, and sequencing
Samples were processed either at the University of Lodz (Poland) or at the Nature Research Centre (Lithuania) (see Supplemental Table SI for details). For samples analyzed at the University of Lodz, total DNA was extracted from the leg of each specimen using the Chelex procedure (see Casquet et al. Citation2012, Morhun et al. Citation2022 for details). For samples analyzed at the Nature Research Center DNA was extracted from the dorsal side using the Quick-DNA Miniprep Plus Kit (Zymo Research) (see Copilaş-Ciocianu et al. Citation2022 for details). The standard animal DNA barcode gene region (COI) (Hebert et al. Citation2003) was amplified using the primers LCO1490, 5′-GGTCAACAAATCATAAAGATATTGG-3′ and HCO2198, 5′- TAAACTTCAGGGTGACCAAAAAATCA-3′ (Folmer et al. Citation1994) with the PCR protocol described by Hou et al. (Citation2007) or with LCO1490-JJ, 5′-CHACWAAYCATAAAGATATYGG-3′ and HCO2198-JJ, 5′-AWACTTCVGGRTGVCCAAARAATCA-3′ (Astrin & Stüben Citation2008) with the PCR protocol described by Copilaş-Ciocianu et al. (Citation2022). PCR products (5 μl) were cleaned up with Exonuclease I (20 U/μl; EURx, Poland) and alkaline phosphatase Fast Polar-BAP (1 U/μl, EURx, Poland) treatment, according to the manufacturer’s guidelines and then sequenced in Macrogen Europe (Netherlands) or BaseClear (Netherlands).
Dataset assembly and phylogeographic analyses
For phylogeographic analysis we included published nucleotide sequences from previous studies (Copilaș-Ciocianu et al. Citation2017; Copilaş-Ciocianu et al. Citation2018; Palatov & Marin Citation2021) in addition to our newly generated sequences. DNA sequences were deposited in the Barcode of Life Data Systems (BOLD - http://v4.boldsystems.org) (Ratnasingham & Hebert Citation2007), in order to obtain the Barcode Index Numbers (BIN) which group DNA sequences based on genetic distance (Ratnasingham & Hebert Citation2013). Relevant voucher information is accessible through the public dataset DS-PCNIPH (DOI: http://dx.doi.org/10.5883/DS-PCNIPH) in BOLD as well as in Supplementary Table SI. Sequence alignment was performed with MUSCLE (Edgar Citation2004) implemented in MEGA 6 (Tamura et al. Citation2013).
Sequence divergences for the COI-5P barcode region (mean and maximum intraspecific variation and minimum genetic distance to the nearest-neighbor species) were calculated using the analytical tools of the BOLD workbench (“Barcode Gap Analysis”), employing the Kimura 2-parameter model (K2P; Kimura Citation1980). We estimated the haplotype number, using the DnaSP v5 software (Librado & Rozas Citation2009). The genetic structure within both species was visualized through a Median-joining network using PopART (Leigh & Bryant Citation2015). Phylogenetic analyses were performed in MEGA 6 (Tamura et al. Citation2013) using the Neighbor-Joining algorithm with Kimura 2-parameter distances, and 1000 bootstrap replicates to visualize relationships among haplotypes.
Results
Field research
In the Dniester basin (Ukraine), N. potamophilus was collected in 2018 from the flooded meadow area beside the river (near Mayaki), in shallow, well-warmed floodplain puddles (both temporary and permanent ones) with almost no water flow (). Amphipods co-occurred alongside A. aquaticus and diverse fauna of freshwater mollusks and leeches. It should be noted that this floodplain meadow has recently been greatly reduced in size due to ongoing urbanization, and the natural floodplain puddles ()) are rare now.
Figure 1. Sampling sites and biotopes where Niphargus hrabei and N. potamophilus were collected: A, B, Dniester River (Ukraine); C, Dnipro River (Ukraine); D, E, Tankovo (Bulgaria); F, Ezerets Lake (Bulgaria).
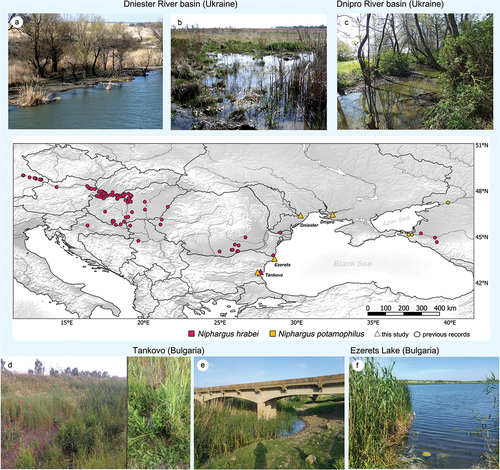
Niphargus potamophilus in the delta of the Dnipro River (Ukraine) was found in 2018 in a shallow stream (near Stara Zburivka) with a lot of decaying vegetation, that flows over an alder forest, in a place where the spring starts its outflow ()). The water in this place was colder than in the surroundings and was flowing slowly. The amphipods co-occurred alongside A. aquaticus and Haemopis sanguisuga (Linnaeus, 1758). This stream is part of the channel that connects the estuarine bay to a system of fish breeding ponds. The amphipods were found in the least anthropogenically transformed part of a former floodplain channel.
In Bulgaria in the Tankovo area, both Niphargus hrabei and N. potamophilus were sampled four years apart. Niphargus hrabei was collected in 2018 from a ditch with apparently temporary water ()). The substrate was muddy with a lot of decaying vegetation. The riparian vegetation was very abundant. The density of the animals was rather high as several tens of individuals were sampled, including 21 females (a few ovigerous), 52 males, and 16 juveniles. The water louse Asellus aquaticus (Linnaeus, 1758) was also highly abundant. This sampling point represents the southernmost record of this species, being situated ca. 150 km south of the previously known locality (Hagieni, Romania; Flot et al. Citation2014). Niphargus potamophilus was sampled in 2014 from Hadjiyska stream, a few tens of meters eastward from where N. hrabei was collected ()). It occurred among reeds on the shallow and muddy shores. Few adult specimens were sampled including both males and females. Accompanying crustacean fauna consisted of Gammarus cf. arduus G. Karaman, 1975, Pontogammarus robustoides (Sars, 1894), and A. aquaticus. This sampling point is the furthest locality from the previously known records (ca. 1000 km) (Palatov & Marin Citation2021).
Niphargus potamophilus in Lake Ezerets (Bulgaria) was sampled in dense aquatic vegetation and muddy substrate along the lake shore in 2018 ()). Only a few immature individuals were sampled that could be properly identified only by DNA barcoding. It was accompanied by typical Ponto-Caspian amphipod fauna such as Pontogammarus robustoides, Dikerogammarus haemobaphes (Eichwald, 1841), Chaetogammarus ischnus (Stebbing, 1899), Trichogammarus trichiatus (Martynov, 1932), Shablogammarus shablensis Carausu, 1943, Lanceogammarus andrussovi (G.O. Sars, 1896), Chelicorophium curvispinum (G. O. Sars, 1895), Ch. sowinskyi (Martynov, 1924), and various unidentified Mysidae.
Morphology of specimens
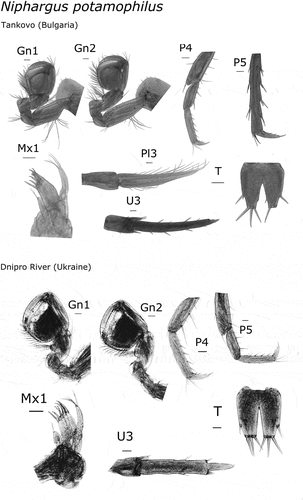
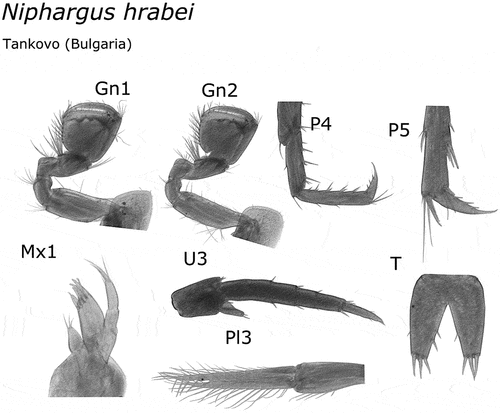
All of the examined morphological characters of our material fell within the range indicated for N. hrabei and N. potamophilus species by Birstein (Citation1954) and the recent redescription of the latter species by Palatov and Marin (Citation2021). Both species are characterized by trapezoidal gnathopod propodi, acutely produced infero-posterior corners of epimeres 2 and 3, additional spines on dactyly of pereopods 3 to 4, uropod 3 non-sexually dimorphic, and the presence of a spoon-shaped appendix on the basis of uropod 1 in males (). The two species can be distinguished from one another by the lack of dorsal spines on telson lobes in N. hrabei (present in N. potamophilus), presence of lateral setae on peduncle of pleopod 3 in N. hrabei (absent in N. potamophilus), inner lobe of maxilla 1 with 3 seate in N. hrabei (one seta in N. potamophilus) (Palatov & Marin Citation2021) (). The morphological identification is in full agreement with the molecular identification through the COI mtDNA barcode.
Molecular studies
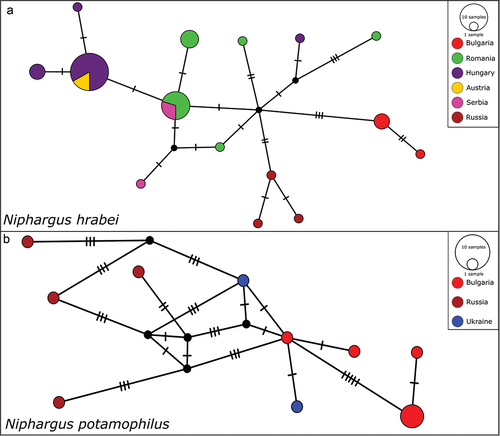
We generated four new sequences for Niphargus hrabei (representing two new haplotypes), and nine new sequences for Niphargus potamophilus (representing six new haplotypes). Analyzing the whole assembled dataset, we identified 15 haplotypes within 48 sequences for N. hrabei, with a 0.41% mean, and 1.31% max intraspecific K2P distance; all sequences were assigned to a single BIN - BOLD:ACQ4737. For N. potamophilus we identified 10 haplotypes within 13 sequences, with a 1.15% mean, and 2.85% max intraspecific K2P distance; all sequences were assigned to a single BIN - BOLD:AEB5889. K2P distance between these two species was 17.25%, clearly indicated in the Neighbor-Joining tree (). The Median-Joining network of N. hrabei shows that individuals from Russia and Bulgaria have only private haplotypes ()). The network for N. potamophilus shows quite different private haplotypes from Russia, with a minimum distance of six substitutions, to the closest haplotypes from Bulgaria and Ukraine ()).
Figure 4. Neighbor-Joining tree depicting the relationship between our newly generated sequences (shown with colored text) and those from previous studies. Numbers at nodes are bootstrap support values (not shown if <60%).
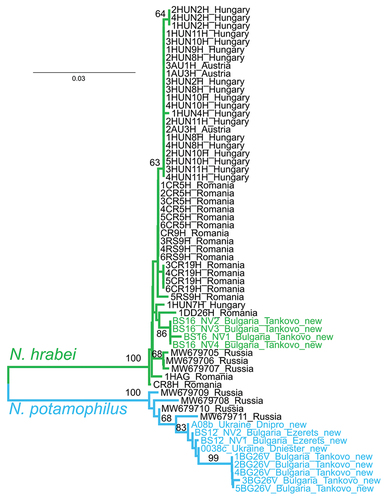
Figure 5. Median-joining network showing the relationships among the haplotypes: a, Niphargus hrabei; b, Niphargus potamophilus. Colors indicate individuals representing different countries of origin. Each bar represents one substitution, whereas small black dots indicate undetected/extinct intermediate haplotype states. The sizes of the circles are proportional to frequencies of haplotypes (see open circles with numbers).
Discussion
The epigean Niphargus species from the Azov-Black Sea region, N. hrabei, N. potamophilus, N. valachicus, and N. magnus Birstein, 1940, have formerly been considered as members of the subgenus Phaenogammarus (Dudich Citation1941) based on their ecological and morphological similarity (Birstein Citation1954; Straškraba Citation1972; Sket Citation1981; Palatov & Marin Citation2021). However, recent molecular research has shown that this similarity is due to convergence, thus rendering Phaenogammarus obsolete (Borko et al. Citation2021). Nevertheless, regarding our focal species, a recent analysis showed that N. potamophilus, N. valachicus, and N. magnus are closely related, forming a well supported clade that is phylogenetically distant from N. hrabei (Palatov & Marin Citation2021). Niphargus hrabei and N. valachicus, are widespread across the middle and lower Danube lowlands and the Black Sea coast (Copilaş-Ciocianu et al. Citation2018; Palatov & Marin Citation2021), while N. magnus and N. potamophilus are known from the eastern Black Sea and Azov-Black Sea basins, respectively (Palatov & Marin Citation2021). Our findings of N. hrabei and N. potamophilus across the west part of the Black Sea basin significantly increased the range of these species, especially for the latter, confirming its westward distribution suggested in the original description (Birstein Citation1954).
The previous and newly gathered ecological data indicates that N. hrabei, N. potamophilus and N. valachicus have similar environmental preferences, occupying a wide range of habitats ranging from springs to small and large streams, large deltaic lakes and channels, estuarine water bodies, and floodplains (Birstein Citation1954; Zorina-Sakharova Citation2017; Copilaş-Ciocianu et al. Citation2018; Palatov & Marin Citation2021; this study). Conversely, N. magnus is distributed both in subterranean and surface water. However, it seems to occupy a narrower spectrum of surface habitats, preferring well-shaded small forest ponds filled with fallen leaves (Birštein Citation1940, Citation1952; Palatov & Marin Citation2021). Such habitats are more isolated as evidenced by the pronounced genetic structuring within this species (Palatov & Marin Citation2021). This is probably the reason why it has a much smaller range than the other three species.
This increase in reports of epigean Niphargus species in the last decade around the Black Sea basin is noteworthy, and could either be a result of more active monitoring, or recent range expansion. The latter in unlikely given the changes in the last century in the condition of large river ecosystems, associated with the simultaneous effects of climate change and anthropogenic flow regulation, which affected the abundance of Ponto-Caspian species and communities (Gogaladze et al. Citation2021), and promoted the spread of non-native species. Furthermore, our molecular analysis showed that Ukrainian and Bulgarian populations of both species are characterized by unique haplotypes, not present anywhere else in the Pannonian or Black Sea basins. As such, it is clear that N. hrabei and N. potamophilus are native species in these areas.
It is, therefore, more likely that these species might have been overlooked previously in the Azov-Black Sea basin, and their dispersal across the marine/brackish waters of the Black Sea might have occurred a long time ago, at the Last Glacial Maximum (30–25 thousand years ago), when this sea was a freshwater lake, isolated from the world ocean (Ryan et al. Citation1997; Bahr et al. Citation2006; Georgievski & Stanev Citation2006). At this time, the Pre-Danube, Pre-Don and Pre-Dnipro paleo-basins were merged into one common alluvial valley (Chepalyga Citation2007; Yanko-Homback et al. Citation2017) and lakes in the Syvash Gulf were desalinated (Olenkovsky Citation2012). This alluvial valley lasted until the latest Mediterranean transgression 16–9 thousand years ago (Yanko-Hombach & Yanina Citation2019: Figure 9). This palaeogeographic context most likely facilitated the dispersal of these freshwater amphipods around the Azov and Black Sea coastline. The phylogeography of both N. hrabei and N. valachicus is consistent with dispersal along the Black Sea coast during this time frame (Copilaş-Ciocianu et al. Citation2018). The subsequent flooding of the Black Sea with saline Mediterranean waters divided the common valley into separate estuaries of the Dnieper, Dniestr, and Danube River basins (Yanko-Homback et al. Citation2017: Figure 16.22), forming the modern Azov and Black Sea (7–9 thousand years ago) (Federov Citation1971; Ryan et al. Citation1997; Badertscher et al. Citation2011; Yanko-Hombach & Yanina Citation2019: Figure 9). Since then, all the major river basins have maintained their isolation. As such, N. hrabei, N. potamophilus and N. valachicus could be present across the entire north Black Sea basin for at least 9 thousand years.
In conclusion, we provide the first morphological and molecular evidence of the presence of N. hrabei in Bulgaria, and N. potamophilus in Bulgaria and Ukraine. Our findings expand the known range of the former ca. 150 km southwards and for the latter species ca. 1000 km westwards. Molecular population structure shows that only Pannonian populations of N. hrabei shared haplotypes between sites and countries, and populations of both species along the Black Sea coast are genetically isolated.
Supplemental Material
Download MS Excel (15.5 KB)Acknowledgements
We thank Andrii Khomenko (V.N. Karazin Kharkiv National University) for providing specimens of N. potamophilus from the Dnipro delta, Andrea Desiderato (University of Lodz) for helping with drafting the map, and Eglė Šidagytė-Copilas (Nature Research Centre, Vilnius) for habitat photographs. The study was supported partially by internal funds of the University of Lodz. HM was supported by the IDUB scholarship of the University of Lodz. DCC was supported by a grant from the Research Council of Lithuania (contract no. 09.3.3-LMT-K-712-19-0149).
Disclosure statement
No potential conflict of interest was reported by the author(s).
Supplementary material
Supplemental data for this article can be accessed online at https://doi.org/10.1080/24750263.2022.2126534
Correction Statement
This article has been republished with minor changes. These changes do not impact the academic content of the article.
References
- Astrin JJ, Stüben PE. 2008. Phylogeny in cryptic weevils: Molecules, morphology and new genera of western Palaearctic Cryptorhynchinae (Coleoptera: Curculionidae). Invertebrate Systematics 22(5):503–522. DOI: 10.1071/IS07057.
- Badertscher S, Fleitmann D, Cheng H, Edwards RL, Göktürk OM, Zumbühl A, Leuenberger M, Tüysüz O. 2011. Pleistocene water intrusions from the Mediterranean and Caspian Seas into the Black Sea. Nature Geoscience 4(4):236–239. DOI: 10.1038/ngeo1106.
- Bahr A, Arz HW, Lamy F, Wefer G. 2006. Late glacial to Holocene paleoenvironmental evolution of the Black Sea, reconstructed with stable oxygen isotope records obtained on ostracod shells. Earth and Planetary Scientific Letters 241(3–4):863–875. DOI: 10.1016/j.epsl.2005.10.036.
- Birstein JA. 1954. Report of epigean Niphargus (Crustacea, Amphipoda) in the Don delta and Kuban basin. Zoologicheskyi Zhurnal 33:1025–1031. In Russian.
- Birštein JA. 1940. To the fauna of cave-dwelling amphipods of Abkhazia. Byulleten Moskovskogo obshchestva ispytatieley prirody. Otdel Biologicheskii 49(3–4):47–55. In Russian.
- Birštein JA. 1952. Subterranean amphipods of the Khosta–Gudauta region (western Transcaucasia). Byulleten Moskovskogo Obshchestva Ispytatieley Prirody. Otdel Biologicheskii 57(1):26–39. In Russian.
- Borko Š, Trontelj P, Seehausen O, Moškrič A, Fišer C. 2021. A subterranean adaptive radiation of amphipods in Europe. Nature Communications 12(1):3688. DOI: 10.1038/s41467-021-24023-w.
- Cărăușu S, Dobreanu E, Manolache C. 1955. Amphipoda. Fauna Republicii Populare Romine. Crustacea 4:92–96.
- Casquet J, Thebaud C, Gillespie RG. 2012. Chelex without boiling, a rapid and easy technique to obtain stable amplifiable DNA from small amounts of ethanol-stored spiders. Molecular Ecology Resources 12(1):136–141. DOI: 10.1111/j.1755-0998.2011.03073.x.
- Chepalyga AL. 2007. The late glacial great flood in the Ponto-Caspian basin. In: Yanko-Hombach V, Gilbert AS, Panin N, Dolukhanov PM, editors. The Black Sea flood question: Changes in coastline, climate and human settlement. Dordrecht: Springer. pp. 119–148.
- Copilaș-Ciocianu D, Fišer C, Borza P, Balázs G, Angyal D, Petrusek A. 2017. Low intraspecific genetic divergence and weak niche differentiation despite wide ranges and extensive sympatry in two epigean Niphargus species (Crustacea: Amphipoda). Zoological Journal of the Linnean Society 181(3):485–499. DOI: 10.1093/zoolinnean/zlw031.
- Copilaş-Ciocianu D, Fišer C, Borza P, Petrusek A. 2018. Is subterranean lifestyle reversible? Independent and recent large-scale dispersal into surface waters by two species of the groundwater amphipod genus Niphargus. Molecular Phylogenetics and Evolution 119:37–49. DOI: 10.1016/j.ympev.2017.10.023.
- Copilaş-Ciocianu D, Grabowski M, Pârvulescu L, Petrusek A. 2014. Zoogeography of epigean freshwater Amphipoda (Crustacea) in Romania: Fragmented distributions and wide altitudinal variability. Zootaxa 3893(2):243–260. DOI: 10.11646/zootaxa.3893.2.5.
- Copilaş-Ciocianu D, Rewicz T, Sands AF, Palatov D, Marin I, Arbačiauskas K, Hebert PDN, Grabowski M, Audzijonyte A. 2022. A DNA barcode reference library for endemic Ponto-Caspian amphipods. Scientific Reports 12(1):11332. DOI: 10.1038/s41598-022-15442-w.
- Dedju I. 1967. Amphipods and mizids of basins of Dniester and Prut rivers. Moscow: Nauka. p. 172. In Russian.
- Dudich E. 1941. Niphargus aus einer Therme von Budapest. Annales Musei Nationalis Hungarici. Pars zoologica 34:165–176. In German.
- Edgar RC. 2004. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Research 32(5):1792–1797. DOI: 10.1093/nar/gkh340.
- Federov PV. 1971. Postglacial transgression of the Black Sea. International Geology Review 14(2):160–164. DOI: 10.1080/00206817209475678.
- Fišer C. 2012. Niphargus: A model system for evolution and ecology. In: Culver DC, White WB, editors. Encyclopedia of caves. New York: Academic Press. pp. 555–564.
- Flot JF, Bauermeister J, Brad T, Hillebrand‐Voiculescu A, Sarbu SM, Dattagupta S. 2014. Niphargus–Thiothrix associations may be widespread in sulphidic groundwater ecosystems: Evidence from southeastern Romania. Molecular Ecology 23(6):1405–1417. DOI: 10.1111/mec.12461.
- Folmer O, Black M, Hoeh W, Lutz R, Vrijenhoek R. 1994. DNA primers for amplification of mitochondrial cytochrome c oxidase subunit I from diverse metazoan invertebrates. Molecular Marine Biology and Biotechnology 3(5):294–299. PMID: 7881515.
- Georgievski G, Stanev EV. 2006. Paleo-evolution of the Black Sea watershed: Sea level and water transport through the Bosporus Straits as an indicator of the Lateglacial-Holocene transition. Climate Dynamics 26(6):631–644. DOI: 10.1007/s00382-006-0123-y.
- Gogaladze A, Son MO, Lattuada M, Anistratenko VV SVL, Pavel AB, Popa OP, Popa LO, ter Poorten J-J, Biesmeijer JC, Raes N, Wilke T, Sands AF, Trichkova T, Hubenov ZK, Vinarski MV, Anistratenko OY, Alexenko TL, Wesselingh FP. 2021. Decline of unique Pontocaspian biodiversity in the Black Sea Basin: A review. Ecology and Evolution 11(19):12923–12947. DOI: 10.1002/ece3.8022.
- Hebert PD, Cywinska A, Ball SL, Dewaard JR. 2003. Biological identifications through DNA barcodes. Proceedings of the Royal Society B: Biological Sciences 270(1512):313–321. DOI: 10.1098/rspb.2002.2218.
- Horton T, Lowry J, De Broyer C, Bellan-Santini D, Coleman CO, Corbari L, Costello MJ, Daneliya M, Dauvin J-C, Fišer C, Gasca R, Grabowski M, Guerra-García JM, Hendrycks E, Hughes L, Jaume D, Jazdzewski K, Kim Y-H, King R, Krapp-Schickel T, LeCroy S, Lörz A-N, Mamos T, Senna AR, Serejo C, Sket B, Souza-Filho JF, Tandberg AH, Thomas JD, Thurston M, Vader W, Väinölä R, Vonk R, White K, Zeidler W. 2022. World Amphipoda Database. DOI: 10.14284/368.
- Hou ZG, Fu JH, Li SQ. 2007. A molecular phylogeny of the genus Gammarus (Crustacea: Amphipoda) based on mito-chondrial and nuclear gene sequences. Molecular Phylogenetics and Evolution 45(2):596–611. DOI: 10.1016/j.ympev.2007.06.006.
- Karaman S. 1950. Etudes sur les Amphipodes-Isopodes des Balkans. Belgrade: Academie Serbe des Sciences Monographies.
- Kimura M. 1980. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. Journal of Molecular Evolution 16(2):111–120. DOI: 10.1007/BF01731581.
- Lefébure T, Douady CJ, Gouy M, Trontelj P, Briolay J, Gibert J. 2006. Phylogeography of a subterranean amphipod reveals cryptic diversity and dynamic evolution in extreme environments. Molecular Ecology 15(7):1797–1806. DOI: 10.1111/j.1365-294X.2006.02888.x.
- Lefébure T, Douady CJ, Malard F, Gibert J. 2007. Testing dispersal and cryptic diversity in a widely distributed groundwater amphipod (Niphargus rhenorhodanensis). Molecular Phylogenetics and Evolution 42(3):676–686. DOI: 10.1016/j.ympev.2006.08.020.
- Leigh JW, Bryant D. 2015. popart: Full-feature software for haplotype network construction. Methods in Ecology and Evolution 6(9):1110–1116. DOI: 10.1111/2041-210X.12410.
- Librado P, Rozas J. 2009. DnaSP v5: A software for comprehensive analysis of DNA polymorphism data. Bioinformatics 25(11):1451–1452. DOI: 10.1093/bioinformatics/btp187.
- Meijering MPD, Jazdzewski K, Kohn J. 1995. Ecotypes of Amphipoda in Central European inland waters. Polskie Archiwum Hydrobiologii 42:527–536.
- Morhun H, Copilas-Ciocianu D, Rewicz T, Son MO, Khomenko A, Huseynov M, Utecsky S, Grabowski M. 2022. Molecular markers and SEM imaging reveal pseudocryptic diversity within the Ponto-Caspian low-profile amphipod invader Dikerogammarus bispinosus. The European Zoological Journal 89(1):87–101. DOI: 10.1080/24750263.2021.2018056.
- Nesemann H, Pöckl M, Wittmann KJ. 1995. Distribution of epigean Malacostraca in the middle and upper Danube (Hungary, Austria, Germany). Miscellanea Zoologica Hungarica 10:49–68.
- Olenkovsky M. 2012. Neolithic of the Sivash region. Kherson: Aylant. p. 176. In Russian.
- Palatov D, Marin I. 2021. Epigean (pond-dwelling) species of the genus Niphargus Schiödte, 1849 (Crustacea: Amphipoda: Niphargidae) from the coastal plains of the Black and Azov seas of the north- and south-western Caucasus. Invertebrate Zoology 18(2):105–151. DOI: 10.15298/invertzool.18.2.05.
- Pérez-Moreno JL, Balázs G, Wilkins B, Herczeg G, Bracken-Grissom HD. 2017. The role of isolation on contrasting phylogeographic patterns in two cave crustaceans. BMC Evolutionary Biology 17(1):247. DOI: 10.1186/s12862-017-1094-9.
- Ratnasingham S, Hebert PD. 2007. BOLD: The Barcode of Life Data System. Molecular Ecology Resources 7:355–364. http://www.barcodinglife.org
- Ratnasingham S, Hebert PD. 2013. A DNA-based registry for all animal species: The Barcode Index Number (BIN) system. PloS ONE 8(7):e66213. DOI: 10.1371/journal.pone.0066213.
- Ryan WBF, Pitman IIIWC, Major CO, Shimkus K, Moskalenko V, Jones GA, Dimitrov P, Gorür N, Sakinç M, Yüce H. 1997. An abrupt drowning of the Black Sea shelf. Marine Geology 138(1–2):119–126. DOI: 10.1016/S0025-3227(97)00007-8.
- Sket B. 1981. Distribution, Ecological, Character and Phylogenetic importance of Niphargus valachicus (Amphipoda, Gammaridae, S.L.). Biology Vestnik 29(1):87–103. In Slovenian.
- Straškraba M. 1972. Les groupments des especes du genre Niphargus (sensu lato). Actes du Ier Colloque International sur le genre Niphargus. Museo Civico di Storia Naturale di Verona Memorie fuori Serie 5:85–90.
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. 2013. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Molecular Biology and Evolution 30(12):2725–2729. DOI: 10.1093/molbev/mst197.
- Trontelj P, Douady C, Fišer C, Gibert J, Š G, Lefébure T, Sket B, Zakšek V. 2009. A molecular test for hidden biodiversity in groundwater: How large are the ranges of macro-stygobionts? Freshwater Biology 54(4):727–744. DOI: 10.1111/j.1365-2427.2007.01877.x.
- Yanko-Hombach V, Yanina T. 2019. Toward an understanding of human responses to environmental change in the Caspian-Black Sea-Mediterranean Corridors (IGCP 610 final report). Episodes Journal of International Geoscience 42(4):343–354.
- Yanko-Homback V, Schnyukov E, Pasynkov A, Sorokin V, Kuprin P, Maslakov N, Montnenko I, Smyntyna O. 2017. Geological and geomorphological factors and marine conditions of the Azov-Black Sea Basin and coastal characteristics as they determine prospecting for seabed prehistoric sites on the continental shelf. In: Flemming NC, Harff J, Moura D, Burgess A, Bailey GN, editors. Submerged landscapes of the European continental shelf: Quaternary Paleoenvironments, first edition. Oxford: Wiley Blackwell. pp. 431–478.
- Zorina-Sakharova K. 2017. Niphargus valachicus Dobreanu et Manolache (Amphipoda, Niphargidae) in Kiliya Delta of the Danube River. Hydrobiological Journal 53(5):46–56. DOI: 10.1615/HydrobJ.v53.i5.50.