Abstract
The mortality of colorectal cancer is expected to increase in some countries including the United States, which necessitates the identification of new molecules that help in prognosis assessment and survival improvement. In this brief report, we evaluated the potential of interleukin-34 (IL-34) as a prognostic factor in colorectal cancer. IL-34 was reported for the first time in 2008 as a novel cytokine that controls the biology of the myeloid cell lineage. Accumulating evidence suggests important roles for IL-34 in modifying the tumor microenvironment and enhancing therapeutic resistance of cancer. In this study, we found that IL-34 expression was detectable in various colorectal cancer cell lines in addition to primary cancer tissues from a cohort of Japanese colorectal cancer patients, ranging from high to absent. A Kaplan–Meier analysis showed that high expression of IL-34 correlated with poor survival of colorectal cancer patients. Importantly, in both univariate and multivariate analysis, high IL-34 expression correlated with unfavorable prognosis. A similar relationship between IL-34 expression and the poorer prognosis was also observed in a cohort of colorectal cancer patients registered at The Cancer Genome Atlas. Together, these findings indicate a potential role for IL-34 as a prognostic factor in colorectal cancer.
Clinical significance
IL-34 expression was detectable in various colorectal cancer cell lines in addition to primary cancer tissues from a cohort of Japanese colorectal cancer patients. A Kaplan–Meier analysis showed that high expression of IL-34 correlated with poor survival of colorectal cancer patients. Importantly, in multivariate analysis, high IL-34 expression significantly correlated with unfavorable prognosis. A similar relationship between IL-34 expression and the poorer prognosis was confirmed in a cohort of colorectal cancer patients registered at The Cancer Genome Atlas. Together, these findings indicate a potential role for IL-34 as a prognostic marker in colorectal cancer.
1. Introduction
Colorectal cancer is the third most frequent cancer worldwide, and the second cause of cancer-related death. According to the World Health Organization (WHO) database (Globocan 2018), colorectal cancer contributes 7.3% (4th rank) and 16.8% (1st rank) of new cases in the United States and Japan, respectively. Furthermore, colorectal cancer showed high mortality rates, with 616714 cases and 409399 cases in the United States and Japan, respectively [Citation1]. Importantly, a recent study by Araghi et al. [Citation2] expected an increase in colorectal cancer mortality rates in some countries by 2035. Based on these backgrounds; prognosis assessment becomes critically important for survival improvement, which can be achieved by evaluating the expression of certain tumor markers and tumor-derived cytokines/chemokines [Citation3,Citation4].
Interleukin-34 (IL-34) is a novel cytokine that was reported for the first time in 2008 as a protein controls the biology of the myeloid cell lineage by the interaction with the colony-stimulating factor-1 receptor (CSF-1R) [Citation5]. The physiological expression of IL-34 is restricted mainly to the brain and skin by neurons and keratinocytes, respectively [Citation6]. In disease, IL-34 expression can be induced by various cells, where it plays pathological roles such as in autoimmune disease, inflammation, infection and metabolic disorders [Citation7]. Accumulating evidence also suggests important roles for IL-34 in cancer, including tumor growth, angiogenesis, metastasis, immunosuppression, therapeutic resistance and pathological osteoclastogenesis [Citation8–19]. In particular, IL-34 contributes to the induction of immunosuppressive tumor microenvironment. IL-34 stimulates CSF-1R that expressed in monocytes and macrophages, then induces them into CD163 positive, immunosuppressive M2-biased tumor-associated macrophages (TAM) [Citation10,Citation11]. It has been known that the CD163 positive TAM contribute to resistance to cancer treatments including anticancer agents and immune checkpoint inhibitors [Citation14]. Consistent with this, the expression of IL-34 in lung cancers correlates adversely with the patient’s survival [Citation11,Citation15].
In this brief study, we evaluate the prognostic value of IL-34 in a cohort of Japanese colorectal cancer patients based on immunohistochemistry staining of IL-34 in cancer tissues. Additionally, we analyzed data from a cohort of colorectal cancer patients registered at large-scale cancer genomics data sets, The Cancer Genome Atlas (TCGA), to evaluate the impact of IL-34 expression on patients’ survival.
2. Materials and methods
2.1. Cell lines
The nine human colorectal cancer cell lines including Caco2, COLO205, HCT116, HT29, LoVo, SW48, SW620, and SW948 cells, and fetal human colon epithelial cells (FHC) were used in this study. All cells were grown in monolayers in the appropriate medium, supplemented with 10% fetal bovine serum (FBS) (Gibco® Tokyo, Japan), and incubated at 37 °C in atmospheres of humidified air with 5% CO2, excepted for SW48, SW620 and SW948 cells with 0% CO2.
2.2. Quantitative real-time PCR (qRT-PCR)
Total RNAs were extracted from cultured cells and clinical tissues using Maxwell® 16 LEV Simply RNA Cells kit and Tissue kit (Promega Co, Madison, WI, USA) according to the manufacturer’s protocol. Complementary DNA was synthesized using ReverTraAce® qPCR RT kit (Toyobo, Osaka, Japan). mRNAs were quantified by real-time PCR analysis using TaqMan® Universal Master Mix II and TaqMan®Gene expression assays on a StepOnePlus (Applied Biosystems, Foster City, CA, USA) according to the manufacturer’s recommendations. Each experiment was done in triplicate. Primers for ACTB (Hs01060665_g1) as internal control and IL-34 (Hs01050926_m1) were used (Applied Biosystems).
2.3. Clinical samples
Colorectal cancer tissues were obtained from patients who had undergone surgery at Kanagawa Cancer Center Hospital (Yokohama, Japan) from 2006 to 2008. A total of 292 resected formalin-fixed paraffin-embedded (FFPE) tumor specimens were preserved at the department of pathology and utilized for immunohistochemical analysis. Other 8 frozen tissue samples for qRT-PCR experiments were obtained with informed consent at Kanagawa Cancer Center Hospital. All tumors were staged based on the pTNM pathologic classification of the UICC (International Union Against Cancer). Clinicopathologic data were shown in . The primary endpoint was overall survival as measured from the date of surgery to the time of death or the last follow-up observation. Written informed consent was obtained from all patients, and the use of related clinical materials was approved by institutional ethics committees, and all experiments were performed in accordance with all guidelines and regulations indicated by these committees.
Table 1. Association of IL-34 protein in colorectal cancer tissues with patients’ characteristics.
2.4. Tissue microarray construction
Tumor tissue microarrays were constructed with 292 formalin-fixed primary colorectal cancers, each of which had been obtained with an identical protocol to collect, fix, and preserve the tissues after resection. The tissue area for sampling was selected based on visual alignment with the corresponding hematoxylin and eosin (H&E)-stained section on a slide. Three to five tissue cores (diameter, 0.6 mm; depth, 3–4 mm) taken from a donor tumor block were placed into a recipient paraffin block with a tissue microarrayer (Beecher Instruments, Sun Prairie, WI, USA). A core of normal tissue was punched from each case, and 5 μm sections of the resulting microarray block were used for immunohistochemical staining of IL-34.
2.5. Evaluation of immunohistochemical staining and statistical analysis
IL-34 positivity was semiquantitatively assessed without prior knowledge of clinicopathologic data. The intensity of IL-34 staining was evaluated by the following criteria: high positive (scored as 2+), dark brown staining in >50% of tumor cells completely obscuring cytoplasm; weak positive (1+), any lesser degree of brown staining appreciable in tumor cell cytoplasm; and absent (scored as 0), no appreciable staining in tumor cells.
2.6. TCGA data
The clinical data and mRNAseq data of colorectal cancer cohort (TCGA, PanCancer Atlas [Citation20]) were obtained from cBioPortal (https://www.cbioportal.org/ [Citation21,Citation22]) and analyzed. Analyses were performed using patient’s vital status in clinical data and patient’s IL34 and CD163 (as M2-biased TAM markers) expression data in mRNAseq data. In the Kaplan–Meier plots, the cut-off point was determined to divide the levels of IL34 and CD163 expression in terms of obtaining the most significant difference within the expression level of 15% to 85%.
2.7. Statistics
Statistical analysis was done using the StatView software. Overall survival curves were calculated from the date of surgery to the time of death or the last follow-up observation. Kaplan–Meier curves were calculated for each relevant variable and IL-34 expression; differences in survival times among patient subgroups were analyzed using the log-rank test. Univariate and multivariate analyses were performed with the Cox proportional hazard regression model to determine associations between clinicopathological variables and cancer-related mortality.
3. Results
3.1. High expression of IL-34 correlates with poor survival in colorectal cancer patients
A previous study by Franzè et al. [Citation13] has described the expression of IL-34 in primary sporadic colorectal cancer tissues. In this study, we started with examining the expression of IL-34 in colorectal cancer cell lines and primary tissues obtained from a cohort of colorectal cancer Japanese patients. It has been reported that normal human colon tissues do not express IL-34 [Citation7]. First, qRT-PCR analysis unveiled enhanced levels of IL34 mRNA transcripts in several human colorectal cancer cell lines including Caco2, CoLo205, SW48, SW480, SW620, SW948 and remarkably in HT29 cell line compared to FHC (). Next, the expression of IL34 was examined in clinical samples from a cohort of colorectal cancer patients. At the mRNA level, the qRT-PCR analysis showed different levels of IL34 mRNA transcripts in primary colorectal cancer tissues compared to the normal colorectal epithelium ().
Figure 1. IL-34 is expressed in colorectal cancer cells lines and primary cancer tissues. (A) qRT-PCR analysis of IL34 expression in colorectal cancer cell lines compared to fetal human colon epithelial cells (FHC). Expression value of each samples were normalized by FHC. Representative data (mean ± SE) of qRT-PCR is shown. Similar results were obtained in two independent experiments. (B) qRT-PCR analysis of IL34 expression in primary colorectal cancer tissues collected from 8 patients (T1, T5, T8, T9, 10, T12, T13 and T14) compared to normal colon epithelium from healthy donor (‘Normal’). Expression value of each samples were normalized by the ‘Normal’ tissue. Representative data (mean ± SE) of qRT-PCR is shown. Similar results were obtained in two independent experiments. (C) Immunohistochemistry staining of IL-34 in primary colorectal cancer tissues compared to the normal human colon epithelium. Representative samples at each expression intensity (high, weak and absent) are shown. (D) A Kaplan–Meier analysis showing overall survival in our cohort of colorectal cancer patients (n = 292) based on IL-34 expression (IL-34High (n = 114), IL-34Weak/Absent (n = 178)).
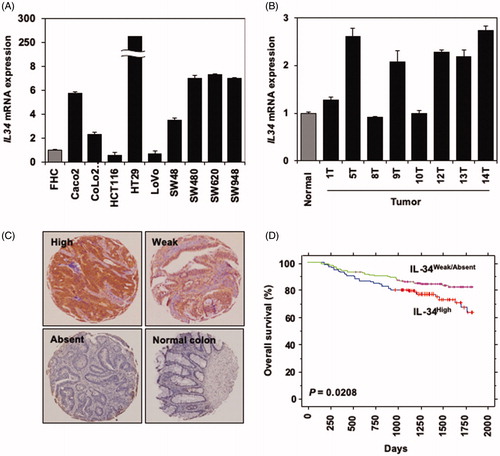
Similarly, IHC staining of clinical samples showed different levels of IL-34 in colorectal cancer tissues ranging from high to absent (). IL-34 staining was not detectable in the normal colon epithelium (). The association of IL-34 staining in colorectal cancer tissues with patients’ characteristics is summarized in . Together, these data indicate that colorectal cancers have the potential to express IL-34, consistent with previous reports [Citation8–19].
Next, we evaluated the impact of IL-34 expression in colorectal cancer tissues on patients’ survival. Among a total of 292 samples in our cohort of Japanese colorectal cancer patients, 114 samples (39.04%) showed strong expression of IL-34 compared to weak (104 samples, 35.61%) or absent (74 samples, 25.34%) (). Based on this categorization, a Kaplan–Meier analysis of overall survival of colorectal cancer patients indicated a significant association between high expression of IL-34 with shorter survival periods compared to other groups that showed weak or absent expression of IL-34 (p = .0208, ). Next, we assessed the association of IL-34 protein expression with the clinical parameters. pT factor (higher in T4; p = .0485 by Fisher’s exact test) was significantly related to high expression of IL-34 (). We also applied univariate analysis to evaluate associations between patient prognosis and other factors, including gender (male vs. female), age (≥65 years vs. <65 years), histologic type (poorly differentiated or mucinous adenocarcinoma vs. well or moderately differentiated adenocarcinoma), pT factor (T4 vs. T1–3), pN factor (N0–1 vs. N2), and IL-34 status (weak, absent vs. high). In a univariate analysis, among these variables, high expression of IL-34 (p = .0227) and advanced pT stage (p < .0001), and advanced pN stage (p < .0001) were significantly associated with poorer prognosis (). In multivariate analysis of the prognostic factors, high IL-34 expression (p = .0331), advanced pT stage (p = .0225), and advanced pN stage (p = .0009) were identified as independent prognostic factors.
Table 2. Cox’s proportional hazards model analysis of prognostic factors in patients with colorectal cancer.
3.2. IL-34 correlates with poor survival in colorectal cancer patients in silico assay
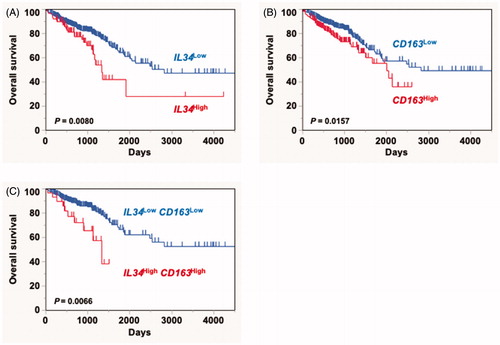
Finally, we utilized the data from a cohort of colorectal cancer patients registered at large-scale cancer genomics data sets, TCGA, through the cBioPortal (www.cbioportal.org) to evaluate the impact of IL-34 expression on patients’ survival. Again, a significant association between high expression of IL-34 with poor survival was observed in the registered cohort compared to other groups (p = .008, ). CD163 serves as a marker for M2-biased immunosuppressive TAM with pro-tumorigenic phenotype. Consistent with previous reports [Citation15], we found that high expression of CD163 correlates with poor prognosis of colorectal cancer patients in this cohort (p = .0157, ). Furthermore, IL-34 was suggested to modify the TAM function at the tumor microenvironment, enhancing its pro-tumorigenic functions such as immunosuppression and metastasis [Citation11,Citation12,Citation14]. Consistent with these backgrounds, Kaplan–Meier analysis of overall survival in colorectal cancer patients based on IL34 and CD163 expression showed that patients with high expression of both IL34 and CD163 have the poorest survival compared to the other group (p = .0066, ). Thus, these findings indicate an association between IL-34 and CD163 expression with poor survival in colorectal cancer.
Figure 2. In silico analysis of the correlation IL34 and CD163 expression with poor survival. A Kaplan–Meier analysis showing overall survival in a cohort of colorectal cancer patients (n = 587) registered at TCGA based on IL34 (IL34high (n = 97), IL34low (n = 490)) (A), CD163 (CD163high (n = 201), CD163low (n = 386)) (B), or co-expression of both molecules (IL34high CD163high (n = 32), IL34low CD163low (n = 321)) (C).
4. Discussion
In this paper, we describe for the first time the prognostic value of IL-34 in colorectal cancer. In a cohort of Japanese colorectal cancer patients, IL-34 expression was detected in cancer tissues at various levels. High expression of IL-34 correlates with the patient’s poor survival. Together, these findings indicate a prognostic value for IL-34 in colorectal cancer and add new evidence to the previous finding that describes the correlation between IL-34 and poor survival in lung cancer [Citation11,Citation15].
Previous reports indicated important pathological roles for IL-34 in diseases of colon and rectum, remarkably in inflammatory bowel disease [Citation19,Citation23]. These studies suggested an enhancement of IL-34 expression at both mRNA and protein levels in the inflamed colon of patients with Crohn’s disease and ulcerative colitis. Furthermore, Franzè et al. [Citation13] described the expression of IL-34 in sporadic colorectal cancer and suggested a role for this cytokine in colon tumorigenesis. Thus, we find that the prognostic value of IL-34 described in this study is consistent with the pathological impact of IL-34 on disease progression, severity and chronicity [Citation6,Citation7].
Colorectal cancer is largely asymptomatic until alarm features develop to advanced stages. Then, it has been recognized that development of sensitive screening system is important to reduce the incidence and mortality rate of colorectal cancer. Currently, various biomarkers have been already clinically used; e.g., fecal haemoglobin, carcinoembryonic antigen (CEA) and CA19-9, although these are not still perfect to predict patients’ prognosis in personalized medicine [Citation24,Citation25]. Therefore, new biomarker(s) which could complement the pre-existing ones for colorectal cancer is expected. Recent advances in cancer biology have indicated a fundamental importance of immunological factor(s) in the occurrence, development and progression of cancer. In this regard, as we have shown in this report, examining IL-34 expression in colorectal cancer tissues, as well as accumulation of CD163 positive macrophages, may help to predict the prognosis of colorectal cancer patients. Particularly, as IL-34 is induicible with inflammatory stimuli, it could be a valuable prognostic marker in colorectal cancer with inflammatory circumstances. However, as we showed IL-34 expression only in the tumor tissues but not in the sera, there could be a practical limitation to use the IL-34 expression as a clinical biomarker. In addition to the pre-existing biomarkers, the IL-34 expression in tumor tissues could be supplementarily used for the management of colorectal cancer patients. On the other hand, IL-34 expressed in colorectal cancer could be a good therapeutic target. Reagents which can inhibit the function or production of IL-34 could be new therapeutic tools for the treatment of colorectal cancer. It is expected that the role of IL-34 in colorectal cancer would be further evaluated in other larger cohorts.
Acknowledgements
The results shown here are in part based upon data generated by the TCGA Research Network (https://www.cancer.gov/tcga).
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Bray F, Ferlay J, Soerjomataram I, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394–424.
- Araghi M, Soerjomataram I, Jenkins M, et al. Global trends in colorectal cancer mortality: projections to the year 2035. Int J Cancer. 2019;144:2992–3000.
- Visconti L, Nelissen K, Deckx L, et al. Prognostic value of circulating cytokines on overall survival and disease-free survival in cancer patients. Biomarkers Med. 2014;8:297–306.
- Kotowicz B, Fuksiewicz M, Jonska-Gmyrek J, et al. The assessment of the prognostic value of tumor markers and cytokines as SCCAg, CYFRA 21.1, IL-6, VEGF and sTNF receptors in patients with squamous cell cervical cancer, particularly with early stage of the disease. Tumor Biol. 2016;37:1271–1278.
- Lin H, Lee E, Hestir K, et al. Discovery of a cytokine and its receptor by functional screening of the extracellular proteome. Science. 2008;320:807–811.
- Baghdadi M, Umeyama Y, Hama N, et al. Interleukin-34, a comprehensive review. J Leukoc Biol. 2018;104:931–951.
- Baghdadi M, Endo H, Tanaka Y, et al. Interleukin 34, from pathogenesis to clinical applications. Cytokine. 2017;99:139–147.
- Baud’huin M, Renault R, Charrier C, et al. Interleukin-34 is expressed by giant cell tumors of bone and plays a key role in RANKL-induced osteoclastogenesis. J Pathol. 2010;221:77–86.
- Cioce M, Canino C, Goparaju C, et al. Autocrine CSF-1R signaling drives mesothelioma chemoresistance via AKT activation. Cell Death Dis. 2014;5:e1167.
- Ségaliny A, Mohamadi A, Dizier B, et al. Interleukin-34 promotes tumor progression and metastatic process in osteosarcoma through induction of angiogenesis and macrophage recruitment. Int J Cancer. 2015;137:73–85.
- Baghdadi M, Wada H, Nakanishi S, et al. Chemotherapy-induced IL34 enhances immunosuppression by tumor-associated macrophages and mediates survival of chemoresistant lung cancer cells. Cancer Res. 2016;76:6030–6042.
- Zhou S, Hu Z, Zhou Z, et al. miR-28-5p-IL-34-macrophage feedback loop modulates hepatocellular carcinoma metastasis. Hepatology. 2016;63:1560–1575.
- Franzè E, Dinallo V, Rizzo A, et al. Interleukin-34 sustains pro-tumorigenic signals in colon cancer tissue. Oncotarget. 2017;9:3432–3445.
- Raggi C, Correnti M, Sica A, et al. Cholangiocarcinoma stem-like subset shapes tumor-initiating niche by educating associated macrophages. J Hepatol. 2017;66:102–115.
- Baghdadi M, Endo H, Takano A, et al. High co-expression of IL-34 and M-CSF correlates with tumor progression and poor survival in lung cancers. Sci Rep.. 2018;8:418.
- Han N, Baghdadi M, Ishikawa K, et al. Enhanced IL-34 expression in Nivolumab-resistant metastatic melanoma. Inflamm Regen. 2018;38:3.
- Baghdadi M, Ishikawa K, Endo H, et al. Enhanced expression of IL-34 in an inflammatory cyst of the submandibular gland: a case report. Inflamm Regen. 2018;38:12.
- Baghdadi M, Ishikawa K, Nakanishi S, et al. A role for IL-34 in osteolytic disease of multiple myeloma. Blood Adv. 2019;3:541–551.
- Franzè E, Monteleone I, Cupi ML, et al. Interleukin-34 sustains inflammatory pathways in the gut. Clin Sci. 2015;129:271–280.
- Hoadley KA, Yau C, Hinoue T, et al. Cell-of-Origin Patterns Dominate the Molecular Classification of 10,000 Tumors from 33 Types of Cancer. Cell. 2018;173:291–304.
- Cerami E, Gao J, Dogrusoz U, et al. The cBio Cancer Genomics Portal: An Open Platform for exploring Multidimensional Cancer Genomics Data. Cancer Discov. 2012;2:401–404.
- Gao J, Aksoy BA, Dogrusoz U, et al. Integrative Analysis of Complex Cancer Genomics and Clinical Profiles Using the cBioPortal. Sci Signal. 2013;6:pl1.
- Zwicker S, Martinez GL, Bosma M, et al. Interleukin 34: a new modulator of human and experimental inflammatory bowel disease. Clin Sci. 2015;129:281–290.
- Yamashita K, Watanabe M. Clinical significance of tumor markers and an emerging perspective on colorectal cancer. Cancer Sci. 2009;100:195–199.
- Sidransky D. Emerging molecular markers of cancer. Nat Rev Cancer. 2002;2:210–219.
