Abstract
Competitive endogenous RNA (ceRNA) plays a vital role in cancer stem cells. In the present study, we aimed to investigate the role of the circular RNA (circRNA) antisense to the cerebellar degeneration-related protein 1 transcript (CDR1as) during competitive inhibition of microRNA-7 (miR-7) in the regulation of autophagy to modulate the proliferation and migration of breast cancer stem cells. The CD44+/CD24- phenotype breast cancer stem cells were sorted from the original MCF7 cell by flow cytometry. RT-qPCR was applied to assess the expression of CDR1as and miR-7 in MCF7-Parent and MCF7-Stem cells. The activity of autophagy was evaluated followed the knockdown and overexpression of CDR1as. The correlation between CDR1as and miR-7 and between CDR1as and miR-7-target mRNAs were evaluated. This study revealed that CDR1as overexpression was associated with up-regulation of autophagy and that CDR1as competitive inhibition of miR-7 enhanced the LKB1-AMPK-mTOR signaling and autophagy-associated genes in breast cancer stem cells. CDR1as promoted the cell growth and migration of breast CSCs via the CDR1as/miR-7 axis, and knockdown of CDR1as and miR-7 overexpression obviously impaired the ability of growth and migration. The present study added a new view of developing a new therapeutic strategy against breast cancer.
Introduction
Breast cancer is the most common malignant tumor among women worldwide and overall the leading cause of cancer-related death in women (Bray et al. Citation2018). Recently, emerging evidences suggest cancer stem cells (CSCs), a subpopulation of cancer cells endowed with self-renewal and differentiation capacity, have been demonstrated to contribute to tumor initiation, progression, treatment resistance and recurrence (Bajaj et al. Citation2020). Therefore, it is necessary to understand the molecular event of breast CSCs underlying progression and to find the novel therapeutic target of breast cancer.
Autophagy is an evolutionally conversed multi-step lysosomal degradation pathway, which could enable tumor cells to survive in nutrient- and oxygen- poor tumor environments (TMEs) (Han et al. Citation2018). Recently, the inhibition of autophagy has been demonstrated to decrease the drug-resistance of breast CSCs and reduce the ability of breast CSCs to form tumors (Yue et al. Citation2013). However, the functional importance and regulation of autophagy remain unclear in breast CSCs.
Circular RNAs (circRNAs), which appear to be as endogenous miRNA sponges since some circRNAs possess microRNA (miRNA) binding sites, allowing them to arrest miRNA activity (Hansen et al. Citation2013). Recently, several circRNAs were found to modulate autophagy activity by regulating the intracellular levels of key autophagy proteins under stress conditions. Antisense to the cerebellar degeneration-related protein 1 transcript (CDR1as) is the most investigated circRNA which contains 63 binding sites for microRNA-7 (miR-7) (Piwecka et al. Citation2017). Knockdown of CDR1as or overexpression of miR-7 promotes the degradation of miR-7-target mRNAs (Chen et al. Citation2020; Xu et al. Citation2018). Recently, our research has confirmed that miR-7 could inhibit autophagy under cancer stringent microenvironment in pancreatic cancer (Gu et al. Citation2017). Thus, we wonder whether the CDR1as/miR-7 signal could affect the level of autophagy in breast CSCs, and further regulate the survival and migration of breast cancer.
In this study, we demonstrated that circRNA CDR1as-induced autophagy regulates the survival and migration of CD44+/CD24- breast cancer stem cells through upregulating autophagy-associated targeted genes of miR-7.
Materials and methods
Cell culture
Breast cancer cell line MCF7 and normal breast epithelial cells MCF10A were purchased from the Cell Bank of Chinese Academy of Science (Shanghai, China). The cells were cultured in DMEM with 10% fetal bovine serum at 37°C in a 5% CO2 humidified incubator. After the cell culture reached 80% confluency, the cells were trypsinized with 0.25% trypsin and harvested. The CD44+/CD24- phenotype breast cancer stem cells were sorted from original MCF7 cells by flow cytometry. The CD44+/CD24-MCF7 breast cancer cells were transfected with EgCDR1as, siCDR1as, EgCDR1as + miR-7 mimics and empty vector for 48 hr or incubated with autophagy inhibitor hydroxychloroquine (HCQ, 25 µM) for 24 hr, then the further experiments were performed.
Flow cytometry assay
The sorting of CD44+/CD24-phenotype was performed by flow cytometry. Briefly, the cells were collected and resuspended in binding buffer and labeled with PE anti-human CD24 antibody and PE-Cy7 anti-human CD44 antibody in the dark at room temperature for 15 min (Antibodies used are listed in Table S1), then sorted using Beckman Coulter MoFlo XDP and analyzed using BD LSRFortessa.
RNA extraction and RT-qPCR
Total RNA was extracted to determine the expression of CDR1as, miR-7, ATG4A, ATG7, and LKB1 by using TRIzol® reagent (Takara Biotechnology Co., Ltd.) according to the manufacturer’s instructions. Reverse transcription reaction was carried out to synthesize complementary DNA by using PrimeScriptTM RT reagent kit (Takara Biotechnology Co., Ltd.). qPCR was performed on the QuantStudioTM 6 Flex system (Applied Biosystems) with the use of the SYBR® Premix Ex TaqTM (Takara Biotechnology Co., Ltd.). The primers for the genes in the present study were synthesized by Sangon (Shanghai, China). All quantitative RT–PCR primer sequences of genes used in the study are listed in Supplemental Table S2.
Cell proliferation assay
The cells were harvested and seeded in 96-well plates at a density of 3×103 cells per well. Cell proliferation was measured using cell counting kit-8 (Dojindo Molecular Technologies, Kumamoto, Japan) for a 2 hr incubation period at 37°C, The absorbance was measured at 450 nm by a microplate reader (Epoch2, BioTek).
Apoptosis assay
The cell apoptosis was analyzed by flow cytometry. The cells were harvested and washed with PBS. An AnnexinV-PE/7-AAD apoptosis detection kit (BD) was used to stain cells at 4°C for 30 min for apoptosis analysis. The data was analyzed using the BD LSRFortessa.
Wound-healing assay
For cell motility assay, the cells were harvested and seeded in 6-well plates to near 80% confluence. A linear wound was carefully made by a 10 μl sterile pipette tip across the confluent cell monolayer. Cell debris was removed by washing with PBS and incubated with DMEM medium. The wounded monolayer was then photographed at 0, 48hr after wounding.
Western blotting
Protein lysates were separated by SDS-PAGE and transferred onto polyvinylidene fluoride membranes (Millipore, USA). The membrane was blocked by 5% skim milk powder for 1 hr, and anti-human LC3, SQSTM1, ATG4A, ATG7, LKB1, pi-LKB1, MAPK, pi-MAPK, mTOR, and pi-mTOR, were added for incubation at 4°C and sequentially hybridized with HRP-labeled secondary antibodies for visualization using ImmobilonTM Western HRP substrate kit (Millipore, USA). The antibodies used are listed in Table S1. Densitometry of specific Western blot bands was analyzed using Quantity One version 4.62 software (Bio-Rad laboratories).
Dual luciferase reporter gene assay
The CDR1as linear sequence, CDR1as-WT was cloned into the pmirGLO vector (Promega, USA), and the sites that may interact with miR-7 were mutated to construct a mutant vector, CDR1as-MT. The miR-7 mimics or mimics-NC were cotransfected with pmirGLO vector into CD44+/CD24- MCF-7 stem cells. The activity of the dual luciferase was detected following the manufacturer’s instructions.
Statistical analysis
Normally distributed data were presented as the mean ± SD of at least three independent experiments. The unpaired t-test was used to compare differences between the two groups. Data were analyzed via one-way ANOVA followed by Dunnett’s post-hoc test for multiple comparisons. Statistical analyses were performed using SPSS 19.0 software (IBM Corp.). All statistical tests were two-tailed, and P<0.05 was considered to indicate a statistically significant difference.
Results
The association between CDR1as and clinical prognosis
To identify the possible relationships between CDR1as and breast cancer in clinical samples, we assessed the prognostic values of CDR1as in breast cancer from The Cancer Genome Atlas (TCGA) database (http://cancergenome.nih.gov/). Although there were no statistically significant differences between the groups in terms of CDR1as expression, we observed that higher CDR1as expression was correlated with the worse patient prognosis in a certain long period of time (Figure A), not only in the luminal type (Figure B) but also in the triple negative breast cancer (Figure C). Previous evidences reported that CD44+/CD24- phenotype cells, as two main signs of the cancer stem cells of breast cancer, were closely associated with a poor prognosis in early invasive breast cancer (Ahmed et al. Citation2012). Thus, we wonder if the CDR1as expression was enhanced in CD44+/CD24- phenotype cells.
Figure 1. CDR1as expression in breast cancer. Correlation analysis between CDR1as and clinical prognosis in breast cancer (A), in luminal type (B) and in triple negative breast cancer (C) from TCGA database. D. The CD44+/CD24-phenotype cells were assessed before and after sorting by flow cytometry. The expressions of CDR1as (E) and miR-7 (F) were determined by qRT-PCR in normal breast cells MCF10A, MCF7-Parent cells and CD44+/CD24-MCF7 breast cancer stem cells. **P<0.01 vs. MCF10A or MCF7-Parent cells.
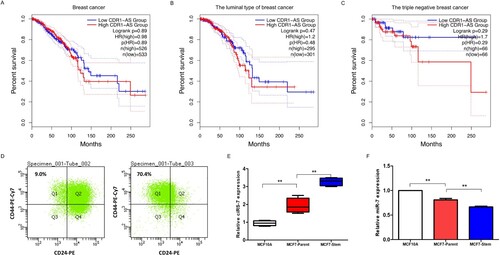
Next, to investigate the expression of CDR1as in breast cancer stem cells, the CD44+/CD24- cells were sorted from the original MCF7 cell by flow cytometry (Figure D). We first explored the expression level of CDR1as in CD44+/CD24- MCF7 breast cancer cells, and the results indicated that the CDR1as level was not only enhanced in breast cancer cells when compared with normal breast cells, but also notably increased in CD44+/CD24- MCF7 breast cancer stem cells when compared with MCF7 breast cancer cells (Figure E). Recently, miR-7 was reported to function as a tumor suppressor and be inhibited by CDR1as in multi-types of cancer (Gu et al. Citation2015), thus, we investigated the expression of miR-7 in breast cancer stem cells. We found the miR-7 was reduced in breast CSCs relative to MCF7 cells (Figure F).
CDR1as promotes but mir-7 inhibits autophagy in breast cancer stem cells
To investigate the role of CDR1as on autophagy in breast cancer stem cells, we firstly evaluate the autophagy activity in CD44+/CD24- tumorigenic breast cancer cells from GSE6883 (http://www.ncbi.nlm.nih.gov/geo). We found breast cancer stem cells increased many autophagy-associated genes when compared with that of normal breast epithelium (Figure A). Thus, we wonder if there’s any relationship between CDR1as and autophagy in CD44+/CD24- breast cancer stem cells.
Figure 2. CDR1as enhanced the autophagy of breast CSCs. A. RNA-seq data of autophagy-associated genes in CD44+/CD24- tumorigenic breast cancer cells obtained from GEO dataset of GSE6883. B. Schematic physical map of recombinant vectors of pLEX-EgCDR1as-CvG2L and pLEX-U6siCDR1as-CvG2L. C. The expressions of CDR1as (left) and miR-7 (right) in CD44+/CD24- MCF7 breast cancer stem cells with knockdown or overexpression of CDR1as. D. The expression of LKB1-AMPK-mTOR pathway in CDR1as-knockdown and -overexpression breast cancer stem cells at 24hr after cultured in 1% FBS for autophagy-induction by Western blot. E. The activity of autophagy was assessed by Western blot after cultured in 1% FBS for autophagy-induction for 24hr.
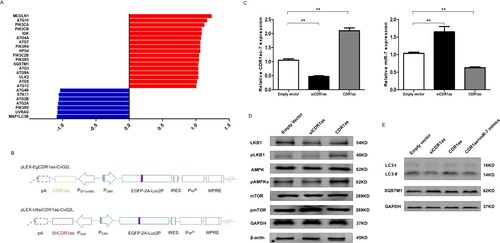
Subsequently, these combinant vectors of pLEX-EgCDR1as-CvG2L and pLEX-U6siCDR1as-CvG2L were constructed (Figure B). The knockdown and overexpression experiments of CDR1as in beast CSCs were performed, and the CDR1as and miR-7 levels were verified (Figure C). To investigate the effects of CDR1as on autophagy, the autophagy-associated signaling was evaluated. pLKB1 and pAMPKα were found to decrease and phosphorylated mTOR increased in breast cancer stem cells with siCDR1as under serum starvation (Figure D). Next, CD44+/CD24- MCF7 breast cancer cell lysates prepared from vector-, siCDR1as, CDR1as, and CDR1as + miR-7 mimics-transfected were analyzed by western blotting for LC3, SQSTM1. We observed increased autophagy in the CDR1as-transfected CD44+/CD24-MCF7 breast cancer cells, whereas it was decreased after transfection with siCDR1as. Moreover, it was found over-expression of miR-7 could reverse the effect of CDR1as on the function of upregulating the autophagy (Figure E). These results suggested that increased CDR1as could accelerate autophagy activity in breast cancer stem cells.
Upregulation of autophagy-associated targeted genes of mir-7 and pathways by CDR1as in breast cancer stem cells
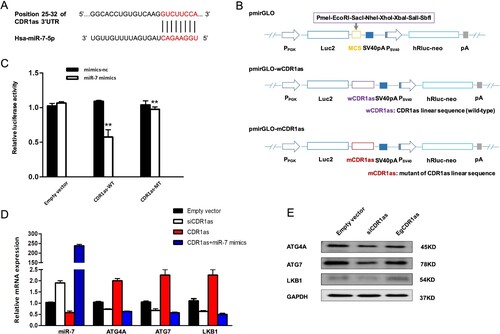
To explore the molecular mechanism of CDR1as on the autophagy, according to the analysis of bioinformatics software TargetScan, it showed that CDR1as has miR-7 binding sites (Figure A). Following, the dual luciferase reporter vectors, including pmirGLO-wCDR1as and pmirGLO-mCDR1as, were constructed (Figure B). The dual luciferase reporter gene assay presented that the miR-7 mimics could decrease luciferase activity in CDR1as-WT groups, but did not affect the luciferase activity in the CDR1as-MT group in CD44+/CD24- MCF7 breast cancer cells (Figure C). Recently, CDR1as has been demonstrated to restrain the availability of miR-7 via modulation of its target genes. Thus, to clarify the potential mechanism of CDR1as-induced autophagy, we analyzed the miR-7 mediated autophagy-associated target genes and pathway under the knockdown of CDR1as in breast CSCs. In this study, we observed that the mRNA (Figure D) expressions, including ATG4A, ATG7 and LKB1, which were identified as miR-7 target genes in our previous study (Gu et al. Citation2017), were remarkably enhanced by CDR1as, but were restored by miR-7 in breast CSCs after treated with miR-7 mimics. Similarly, CDR1as-knockdown markedly decreased the target-gene protein levels, whereas protein levels increased by overexpression of CDR1as (Figure E). These data suggested that CDR1as might activate autophagy via target genes of miR-7.
Figure 3. The molecular mechanism of CDR1as in the activation of autophagy. A. Prediction of binding sites between CDR1as and miR-7 by TargetScan. B. The dual luciferase reporter vectors, pmirGLO-wCDR1as and pmirGLO-mCDR1as, were constructed. C. Identification of the interaction between CDR1as and miR-7 by the dual luciferase reporter gene assay. D-E. Expression of ATG4A, ATG7, and LKB1 in CD44+/CD24-MCF7 breast cancer cells via RT-qPCR analysis (D) and western blotting (E).
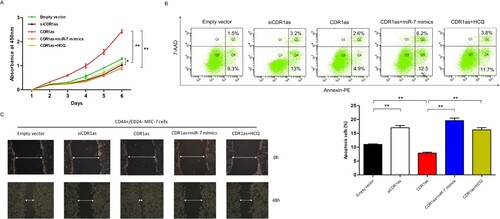
CDR1as promotes the cell growth and migration of breast CSCs via mir-7
To confirm the effect of CDR1as on tumorigenic and invasion role, CD44+/CD24- MCF7 breast cancer cells were transfected with siCDR1as, EgCDR1as with or without miR-7 mimics, we found that knockdown of CDR1as could inhibit the cell vitality of breast CSCs, and the miR-7 mimics or autophagy inhibitor HCQ abrogated the effects of CDR1as and restored the cell vitality (Figure A). Meanwhile, the apoptosis was determined by flow cytometry. The results showed that knockdown of CDR1as promoted apoptosis but overexpression of CDR1as inhibited apoptosis, which could be recovered by miR-7 mimics or HCQ in breast CSCs (Figure B). Moreover, we found that knockdown of CDR1as and miR-7 overexpression obviously impaired the ability of migration, and the miR-7 mimics or repressing the autophagy could recover CDR1as induced migration (Figure C). These findings suggested that CDR1as could promote tumor growth and migration through autophagy-associated target genes of miR-7 in breast CSCs.
Figure 4. CDR1as accelerates breast cancer stem cell growth and metastasis. A. Cell proliferation of breast cancer stem cells was assessed by CCK8 assay (n = 3/group). B. Flow cytometry was used to analyze the apoptosis in breast cancer stem cells when they were transfected with either siCDR1as, CDR1as, or CDR1as combined with miR-7 mimics after 48hr or with HCQ after 24hr. (**, P<0.01) C. Cell motilities of breast cancer stem cells transfected with siCDR1as, CDR1as, or CDR1as combined with miR-7 or HCQ were observed at 0 and 48hr following wounding by a pipette tip.
Discussion
Autophagy, which could be induced by cancer therapy or other nutrient stresses, frequently contributes to cancer cell survival through the recycling of their constituent metabolites (Smith and Macleod Citation2019). Recently, cancer stem cells were reported to represent protective autophagy and efficient cell cycling, along with highly qualified epithelial–mesenchymal transition (EMT) regulators and anti-apoptotic and DNA repairing systems (Najafi et al. Citation2019). Autophagy is one pivotal process that is strongly associated with CSCs maintenance and aggressiveness. Moreover, increasing evidences demonstrated breast cancer stem cells are responsible for tumor recurrence and metastasis, thus affecting the treatment progress (Dittmer Citation2018). Hereinabove, we reported a high level of autophagy is detected in breast cancer stem cells. Further, down-regulation of CDR1as with siCDR1as in breast cancer stem cells inhibited the level of autophagy activity. While repressing the autophagy, breast cancer stem cells overexpressing CDR1as were unable to proliferate exponentially and migration as expected. In other words, the de-activation of autophagy decreased the effects of CDR1as on breast cancer stem cells. Thus, these results suggested that CDR1as could regulate the survival and migration of breast cancer stem cells through activating autophagy.
CircRNAs, a special group of long-chain and non-coding RNAs (ncRNAs), widely exist in mammalian cells and plays a role in regulating gene expression, especially as works competitive endogenous RNAs (ceRNAs) (Patop et al. Citation2019). Recently, studies have demonstrated that circRNAs act as miRNA sponges to regulate the function of miRNAs. Increasing evidences indicates that circRNAs and targeted miRNAs are involved in the development, progression and metastasis of various cancers, such as liver cancer, bladder cancer, esophageal cancer, ovarian cancer, and other cancers (Kristensen et al. Citation2018). Currently, CDR1as has been proved to act as a ceRNA to competitively bind to miR-7 and thus inhibits its activity (Zou et al. Citation2019). In this study, we found that CDR1as is a highly expressed circular RNA in breast cancer stem cells. Moreover, CDR1as negatively correlated to the expression of miR-7. Overexpression of CDR1as not only stimulated breast cancer stem cells autophagy, but also increased cell proliferation and survival, which could be abrogated by knockdown of CDR1as or overexpression miR-7 to induce apoptosis. Furthermore, we observed that inhibition of autophagy with siCDR1as was able to impair migration. These suggested that CDR1as/miR-7 axis affects the biological behavior of breast cancer stem cells.
Then, the specific molecular mechanism of CDR1as/miR-7 in the regulation of autophagy in breast cancer stem cells was further investigated. We observed that CDR1as-7/miR-7 modulated distinct steps of autophagy in breast cancer stem cells by targeting autophagy-related genes, including LKB1, ATG4A and ATG7, which act at distinct steps of autophagy (Bernard et al. Citation2020). Tumor suppressor gene LKB1 serves as the upstream molecule controlling AMPK activation. AMPK (adenosine monophosphate-activated protein kinase), one of the most important molecular energy sensors, regulates autophagic degradation and adapts it to cellular energetic status. AMPK activation could lead to inhibit its downstream signal mTOR cascade, which serves as the main regulator of autophagy. Under stressful conditions, mTOR suppression triggers the autophagic cascade and inhibits the cell proliferation (Li and Chen Citation2019). In breast cancer stem cells, we found that overexpression of CDR1as upregulated the targets of miR-7 LKB1, following activating AMPKα and repressing mTOR signaling, and enhancing the autophagy. Recent findings also suggested that AMPK was critical in modulating the self-renewal and chemoresistance of cancer stem cells (Wang et al. Citation2016).
In brief, this study demonstrated that CDR1as overexpression is associated with up-regulation of autophagy and that CDR1as competitive inhibition of miR-7 enhanced the LKB1-AMPK-mTOR signaling and autophagy-associated genes in breast cancer stem cells. In addition, knockdown of CDR1as or overexpression miR-7could regulate the proliferation and migration of CD44+/CD24- phenotype breast cancer stem cells in vitro. Based on these observations, it suggested that the aberrant CDR1as/miR-7 axis could serve as a promising target for therapeutic potentials in breast cancer.
Supplemental Material
Download MS Word (18.4 KB)Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement (DAS)
Because the main research has not been completed, participants of this study did not agree for their data to be shared publicly.
Additional information
Funding
References
- Ahmed MA, Aleskandarany MA, Rakha EA, et al. 2012. A CD44(-)/CD24(+) phenotype is a poor prognostic marker in early invasive breast cancer. Breast Cancer Res Treat. 133:979–995.
- Bajaj J, Diaz E, Reya T. 2020. Stem cells in cancer initiation and progression. J Cell Biol. 219:e201911053.
- Bernard M, Yang B, Migneault F, et al. 2020. Autophagy drives fibroblast senescence through MTORC2 regulation. Autophagy. 16:2004–2016.
- Bray F, Ferlay J, Soerjomataram I, et al. 2018. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 68:394–424.
- Chen L, Shi J, Wu Y, et al. 2020. CircRNA CDR1as promotes hepatoblastoma proliferation and stemness by acting as a miR-7-5p sponge to upregulate KLF4 expression. Aging (Albany NY). 12:19233–19253.
- Dittmer J. 2018. Breast cancer stem cells: features, key drivers and treatment options. Semin Cancer Biol. 53:59–74.
- Gu DN, Huang Q, Tian L. 2015. The molecular mechanisms and therapeutic potential of microRNA-7 in cancer. Expert Opin Ther Targets. 19:415–426.
- Gu DN, Jiang MJ, Mei Z, et al. 2017. microRNA-7 impairs autophagy-derived pools of glucose to suppress pancreatic cancer progression. Cancer Lett. 400:69–78.
- Han Y, Fan S, Qin T, et al. 2018. Role of autophagy in breast cancer and breast cancer stem cells (review). Int J Oncol. 52:1057–1070.
- Hansen TB, Jensen TI, Clausen BH, et al. 2013. Natural RNA circles function as efficient microRNA sponges. Nature. 495:384–388.
- Kristensen LS, Hansen TB, Veno MT, et al. 2018. Circular RNAs in cancer: opportunities and challenges in the field. Oncogene. 37:555–565.
- Li Y, Chen Y. AMPK and autophagy. Adv Exp Med Biol 2019;1206:85-108.
- Najafi M, Mortezaee K, Majidpoor J. 2019. Cancer stem cell (CSC) resistance drivers. Life Sci. 234:116781.
- Patop IL, Wust S, Kadener S. 2019. Past, present, and future of circRNAs. EMBO J. 38:e100836.
- Piwecka M, Glazar P, Hernandez-Miranda LR, et al. 2017. Loss of a mammalian circular RNA locus causes miRNA deregulation and affects brain function. Science. 357:eaam8526.
- Smith AG, Macleod KF. 2019. Autophagy, cancer stem cells and drug resistance. J Pathol. 247:708–718.
- Wang Z, Wang N, Liu P, et al. 2016. AMPK and cancer. Exp Suppl. 107:203–226.
- Xu B, Yang T, Wang Z, et al. 2018. CircRNA CDR1as/miR-7 signals promote tumor growth of osteosarcoma with a potential therapeutic and diagnostic value. Cancer Manag Res. 10:4871–4880.
- Yue W, Hamai A, Tonelli G, et al. 2013. Inhibition of the autophagic flux by salinomycin in breast cancer stem-like/progenitor cells interferes with their maintenance. Autophagy. 9:714–729.
- Zou Y, Zheng S, Deng X, et al. 2019. The role of Circular RNA CDR1as/ciRS-7 in regulating tumor microenvironment: A Pan-cancer analysis. Biomolecules. 9:429.
