?Mathematical formulae have been encoded as MathML and are displayed in this HTML version using MathJax in order to improve their display. Uncheck the box to turn MathJax off. This feature requires Javascript. Click on a formula to zoom.
?Mathematical formulae have been encoded as MathML and are displayed in this HTML version using MathJax in order to improve their display. Uncheck the box to turn MathJax off. This feature requires Javascript. Click on a formula to zoom.Abstract
Stemness is now an accepted cause of treatment failure and relapse in cancer patients. Laryngeal squamous cell carcinoma patients face a high recurrence and metastatic disease. Research efforts have focused in recent years on discovering biomarkers of poor prognosis in laryngeal cancer patients. SALL4 is an embryonic stem cell factor which has been reported to be highly expressed; and to play a role in oncogenesis and progression in several malignant tumours. As of yet, the expression of this protein has not been assessed in laryngeal cancer. This study provides evidence of positive expression of this protein in laryngeal cancer cells, with positive correlation in the same tumour cells with a second stemness agent, CD 44. Furthermore, it discusses the shared pathways between the two proteins in EMT, Wnt pathway and stemness phenomenon; and proposes further studies to elucidate the role of this protein in laryngeal squamous cell carcinoma.
Introduction
Despite progress in comprehensive treatment regimens, laryngeal cancer patients still face a 30–40% chance of tumour recurrence or metastases (Yu et al. Citation2013). Aggressive surgical, adjuvant and gene therapies have resulted in a higher 5-year survival rates for patients with this type of cancer, but failed to stop metastases and recurrence (Yu et al. Citation2013). One of the causes of failures of conventional cancer treatment is inter-tumoral heterogeneity (Dawood et al. Citation2014). Presence of cancer stem cells is now an accepted important cause of this intra-tumoral heterogeneity (Dawood et al. Citation2014). Cancer stem cells (CSC) is a subpopulation of cells within a tumour which appears to have an exclusive capability of initiating tumorous growth, for self-renewal and to differentiate into terminal cancer cells (Janisiewicz et al. Citation2012). While their presence was disputed for a long period of time, experimental evidence of the existence of cancer stem cells was confirmed in several cancer subtypes, including acute myelogenous leukaemia, breast, brain, prostate, colon and lung cancers (Dawood et al. Citation2014).
The study of CSC in laryngeal cancer started recently (Yu et al. Citation2013). Identifying these CSCs and charting their mechanisms of actions is of prime importance in the management of cancer (Barbato et al. Citation2019). Since stem cells are not present in most adult tissues, they could be ideal targets for cancer-specific diagnosis and treatment (Tatetsu et al. Citation2016).
Recently, Spalt-Like Transcription Factor 4 (SALL4) was reported to be a marker of cancer stem cells. This zinc-finger protein has a unique role in connecting the dots among embryonic stem cells, developmental biology and cancer. By interacting with various epigenetic modulators and sequence-specific transcription factors, it exerts downstream target gene expression, thus making it a key cell fate regulator (Tatetsu et al. Citation2016).
SALL4 plays an essential role during embryonal development, forming a regulatory network with other stemness-related genes, such as the Octamer-Binding Transcription Factor 4 (OCT-4); the Nanong Homebox (NANOG) and the Sex-Determining Region Y-Box 2 (SOX2) (Cheng et al. Citation2016).
By acquiring stemness-related phenotype, through the expression of SALL4 and co-stemness related genes, the cancer cell gains the ability to move freely, invade and generate its own vascular network, develop resistance to apoptosis, acquire the ability to resist chemotherapy, and finally develop the ability to evade the immune system (Zhang et al. Citation2015; Cheng et al. Citation2016; Nicolè et al. Citation2017). In a recent metanalysis analysing the association of SALL4 with pathological, demographical and clinical aspects of cancers, SALL4 expression correlated with increased rates of recurrence and shortened survival (Nicolè et al. Citation2017).
Another intriguing aspect of SALL4 role in tumorigenesis involves its relationship with long non-coding RNAs (lncRNAs) molecules (Ma et al. Citation2019). lncRNA are non-coding transcripts; transcribed by RNA polymerase; without having the ability to code for protein synthesis (Si et al. Citation2016). They participate in diverse biological processes including chromatin remodelling, transcriptional and post-transcriptional regulation (Qi et al. Citation2017). Studies indicate that an increasing number of lncRNAs are associated with stem cell phenotype of CSCs, such as the lncRNA ROR, which by sponging many miRNAs, it acts to modulate the stemness of tumour cells (Hou et al. Citation2014; Gao et al. Citation2016; Fu et al. Citation2017).
SALL4 appears to have a close relationship with lncRNAs. Yuan JH and co-workers demonstrated that SNHG12, an lncRNA, increases proliferation, migration and invasion; and represses apoptosis of breast cancer cells by upregulating SALL4 expression, through its sponging of the miR-15a-5p (Yuan et al. Citation2020). On the other hand, ‘differentiation antagonizing non-protein coding RNA’ (DANCR), which is another lncRNA, was shown to be upregulated by SALL4 in gastric cancer cells. DANCR knockdown inhibited the migration and invasion of gastric cells, while its over-expression had the opposite effects (Pan et al. Citation2017). In both breast and gastric cancers, dual-luciferase reporter assay confirmed the interaction of the respective lncRNA with SALL4 (Pan et al. Citation2017; Yuan et al. Citation2020).
SALL4 expression is silenced in adult, differentiated tissues (Tatetsu et al. Citation2016). Evidence suggests that a recurrence of SALL4 expression later in adulthood could be associated with neoplastic transformation in different sites (Nicolè et al. Citation2017). Review of the literature provides evidence of SALL4 heavy involvement in various cancers such as leukaemia, germ cell tumours, liver cancer, gastric cancer, colorectal cancer, oesophageal cancer, breast cancer, endometrial cancer, lung cancer and glioma (Tatetsu et al. Citation2016 Jun 15). From a functional point of view, the SALL4 gene is important for survival (Ueno et al. Citation2014), drug resistance (Li et al. Citation2015) and metastasis (Zhang et al. Citation2014) of different types of cancer cells.
This protein is also recognized as a potential biomarker for assessing course of the disease. For example, a positive SALL4 expression is strongly correlated with increased overall cancer death and recurrence (Nicolè et al. Citation2017; Shen et al. Citation2017). Knocking-down the expression of SALL4 resulted in the inhibition of tumour’s growth in xenograft models of hepatocellular carcinoma (Yong et al. Citation2013). Levels of expressions of this protein in the serum and tissues correlated with lymph node metastases, histological grade and clinical stage of colon cancer (Wu et al. Citation2017). Knocking-down SALL4 expression resulted in sensitizing nasopharyngeal tumour cells to radiation, increased radiation-induced DNA damage, increased apoptosis of tumour cells and tumour cell cycle arrest (Nie et al. Citation2019). In haematological malignancies, SALL4 high expression is associated with high-risk myelodysplastic syndromes with poor survival (Wang et al. Citation2013). In germ cell tumours, SALL4 is a sensitive diagnostic marker (Mei et al. Citation2009). Furthermore, an anti-SALL4 antibody targeting circulating germ cell tumour cells has shown more sensitivity than α-fetoprotein and glypican (Nastaly et al. Citation2014).
CD44, on the other hand, which is among the most frequent markers of CSCs in cancer, has been shown to be the target of the Wnt pathway (Kwong and Dove Citation2009). CD44 is a glycoprotein located on the cell surface, functioning as a major receptor of hyaluronic acid, which is involved in cell cohesion and migration (Gao et al. Citation2011; Masuko et al. Citation2012). In head and neck cancer, cells over-expressing CD44 were shown to result in higher proliferative ability than cells lacking this expression (Kokko et al. Citation2011; Chai et al. Citation2014; Shen et al. Citation2014; Greco et al. Citation2016). This over-expression was also accompanied with poor 5-year survival rates in patients with squamous cell laryngeal tumours with high expression of this marker (Kokko et al. Citation2011; Shen et al. Citation2014). Other studies found that CD44 expression may predict the outcome of radiotherapy in head and neck cancer patients by assessment of CSC density, where CD44 was the only marker with significant correlation with clinical response to radiotherapy in early-stage LSCC patients (Baumann and Krause Citation2010; de Jong et al. Citation2010).
SALL4 protein expression has not been studied yet in laryngeal cancer. This study will analyse the expression of SALL4 in LSCCs, correlating this expression with clinical-pathological attributes of the tumour cells; and with CD44 expression.
Materials and methods
Ethics statement
The study was conducted in accordance with the Declaration of Helsinki, and approved by the Institutional Review Board (IRB) of the College of Medicine at Alfaisal University.
Tissues
Two commercial human larynx microarrays (Catalogue no. LP803; Biomax US, Rockville, MD), with 80 cases of laryngeal squamous cell carcinomas each, were used for this study. All the cases were confirmed laryngeal squamous cell carcinomas. Major parameters of these tumours include age, sex, anatomic site, pathologic diagnosis and grade, clinical stage and TNM staging. For comparison with normal laryngeal mucosa, a second microarray (Catalogue no. FDA 992, Biomax US, Rockville, MD) carrying normal tissues from all human anatomic sites was used.
Immunohistochemistry (IHC)
For immunostaining, the slides were deparaffinized and epitopes were retrieved using Dako Retrieval Solution (Dako Cytomation, USA) at 95 C for 30 min, followed by cooling to room temperature for 2 min. 0.3% H2O2 was used to inactivate endogenous peroxidase. Subsequently, the sections were rinsed twice in PBS for 5 min (Ouban and Ahmed Citation2015).
Immunostaining was done with antibodies directed against SALL4 (rabbit, polyclonal, ab 29112, UK), and CD 44 (rabbit, polyclonal, ab 157107, abcam, UK); following dilution in antibody diluent (Agilent, Santa Clara, California). The Vectastatin ABC peroxidase kit was used according to the manufacturer’s instructions (Vectastatin Elite ABC Kit, Vector Laboratories, USA). Negative controls were used with omission of primary antibody. Separate positive controls of mixed tumour (SALL4) and tonsils (CD44) were used for test optimization and run validation.
Staining evaluation
Immunohistochemical staining of all tumours was evaluated by two pathologists (AO) and (ER), independently, and a consensus was reached for discordant cases. The degree of SALL4 and CD44 reactivity was scored by applying a semi-quantitative immunoreactivity scoring (IRS) system as described by Baccelli and co-workers (Baccelli et al. Citation2014).
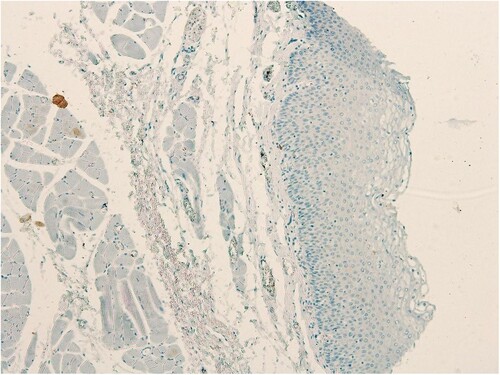
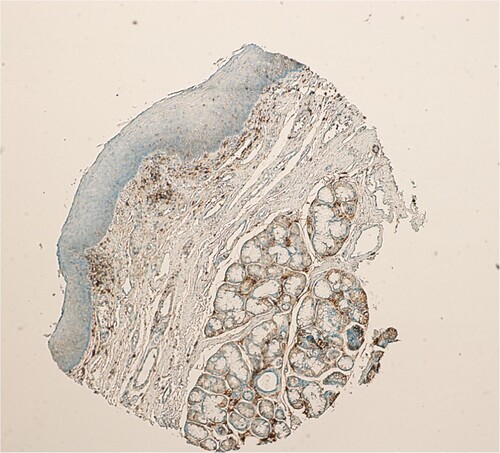
The staining intensity was categorized into four grades: 0, no immunostaining; 1, weak staining (light yellow); 2, moderate staining (buffy); and 3, strong staining (brown). The percentage of positive cells was categorized into five grades: 0 (0%); 1, (1–10%); 2 (11–50%); 3 (51–80%); and 4 (>80%). The staining intensity and percentage of positive cells were multiplied to obtain an IRS for SALL4 expression, in the range of 0–12 for each individual case (Baccelli et al. Citation2014). SALL4 and CD44 expressions in normal, benign, unmatched laryngeal mucosa (Catalogue no. FDA 992, Biomax US, Rockville, MD), were both negative (Figure and Figure , respectively). In line with studies of the immunostains performed on other tissues; an LSCC case was scored as positive for SALL4 and CD44, with an IRS of ≥3 defined as positive expression (Hao et al. Citation2016; Di et al. Citation2018).
Statistical analysis
Immuno-expression of SALL4 was correlated with parameters such as gender, age, differentiation grade, clinical stage and TNM staging. The association of the immune-expressions between SALL4 and CD 44 was assessed for statistical significance. All statistical analyses were performed with IBM SPSS Statistics software package, version 25.0. p < 0.05 was considered to indicate a statistically significant difference. Chi-squared test and Fisher’s exact test were used to test significance.
Results
SALL4 is positively expressed in LSCCs; and the majority of the tumours express SALL4 in a cytoplasmic/nuclear pattern
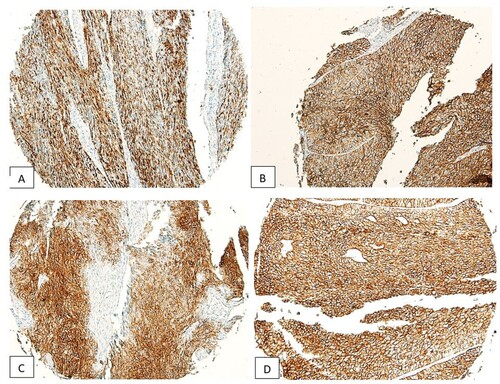
The human larynx microarray (Catalogue no. LP803; Biomax US, Rockville, MD) with 80 cases of laryngeal squamous cell carcinomas was used for this study. The age of the cancer patients in this array was in the range of 39–72 years, with a mean of 53 years. LSCC grades were distributed as follows: 25 grade I; 44 grade II and 5 were grade III. Six tumours in the microarray did not have a grade designation reported by the manufacturer of the array. The expression of SALL4 in normal, benign, unmatched laryngeal mucosa (Catalogue no. FDA 992, Biomax US, Rockville, MD) was absent (IRS# 0–2, Figure ). In laryngeal SCC cases, on the other hand, 57/80 cases were positive for SALL4 (71.25%, χ2 = 9.831; p < 0.001, Table , Figure (A–D). The rest of the cases (23 in total), had weak or negative SALL4 expression (IRS#0-2). A significant majority of SALL4-positive laryngeal SCC cases (53/57, 92.9%, p < 0.001) expressed SALL4 in a combined pattern in the cytoplasm and nuclei of the tumour cells (Figure (A–D), Table ). There was no statistically significant association of immunoexpression of SALL4 and age (p = 0.315), gender (p = 0.711), histological grade (p = 0.618), clinical stage (p = 0.484), T stage (p = 0.324), N stage (p = 0.524) and M staging (p = 1) (all data in Table ).
Table 1. SALL4 expression in LSCC cases and its subcellular localization.
Table 2. SALL4 protein expression status in laryngeal squamous cell carcinomas to assess the relationship between SALL4 positivity and age, gender, grade, clinical stage, T stage, N stage and M staging.
CD44 is highly expressed in LSCCs tumours
The expression of CD44 in normal, benign, unmatched laryngeal mucosa (FDA 992, Biomax) was negative (Figure ). On the other hand, in LSCC cases, 65/80 cases were positive for CD44 (81.25%, χ2 = 21.779; p < 0.001, Table , Figure (A–D)).
Figure 4. A-D, CD44 Positive Cases, Heavy Nuclear and Cytoplasmic Expression. Figure (C) Shows an Embolus of LSCC Tumour Cells Expressing the CD44.
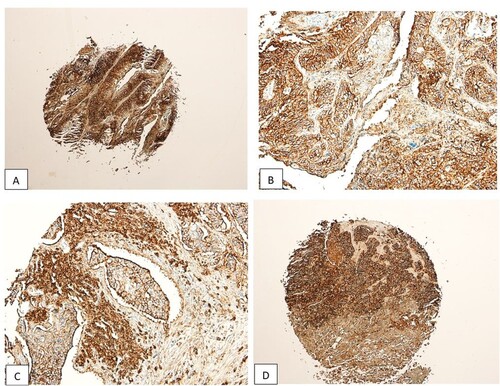
Table 3. CD44 expression in laryngeal squamous cell cancers.
Association between SALL4 and CD 44
Among the LSCC cases, 65 cases were positive for CD 44 immunostain. Fifty of 57 (50/57) SALL4 positive cases were also positive for CD 44 (χ2 = 23.085, p < 0.001). Seven cases were positive for SALL4 and negative for CD 44 (Table ).
Table 4. Assessment of the association between SALL4 and CD44 in LSCC tumour cells.
Discussion
This is one of the first studies analysing the expression of SALL4 in LSCCs, and the first to assess SALL4 and CD44 expressions in the same LSCCs. This study shows that SALL4 expression is significant in LSCCs (p < 0.001). The SALL4 marker was significantly localized in the cytoplasmic/nuclear regions within the LSCC cells (p < 0.001). On the other hand, CD 44 was also positively expressed in LSCC patients (p < 0.001). Furthermore, strong correlation is seen between the two markers in the same tumour cells (χ2 = 23.085, p < 0.001). Collectively, these results may indicate a strong stemness role in the development and maintenance of neoplastic cells in LSCCs. While no association was found between SALL4 expression and clinical and pathological parameters, this may be due to the possibility that stemness and SALL4 expression are late events in tumorigenesis, mostly affecting survival, recurrence and chemotherapy resistance (Ueno et al. Citation2014; Zhang et al. Citation2014; Li et al. Citation2015), attributes which were lacking in the data provided. Given the heterogeneity seen in LSCC tumour biology, even for patients presenting with similar sites and stages (Baumann and Krause Citation2010), this study may shed some light on this phenomenon; and may provide future directions of research addressing the stemness phenomenon in LSCC patients.
The mechanism driving the re-activation of SALL4 in cancer remains largely unknown, however there are several hypotheses. Du and co-workers in 2018, reported a dramatic increase in SALL4 mRNA and protein levels 24-hours following treatment of a lung cancer cell line with Epidermal Growth Factor (EGF) (Du et al. Citation2018 Apr Citation25). Treatment of a second lung cancer cell line with ERK inhibitor resulted in a steep knockdown of SALL4 expression. This showed that ERK
signalling may be involved in SALL4 regulation by EGFR (Du et al. Citation2018 Apr Citation25). Interestingly, and in line with the results of this current study; the expression of SALL4 correlated with that of CD44 in the same non-small cell lung cancer cells (Du et al. Citation2018 Apr Citation25). A second study reported that the canonical WNT signalling may directly regulate expression of SALL4, and that this regulation is achieved by direct interaction of the Lymphoid-Enhancer-binding-factor-1 (LEF-1) (Böhm et al. Citation2006). SALL4 was also identified as a direct target of Caudal-related homebox (CDX1) protein (Fujii et al. Citation2012). In murine embryonic stem cells, several studies reported that SALL4 protein participates in an interconnected autoregulatory circuit with Oct4, Sox2 and Nanog, wherein each of the four factors may regulate its own expression; as well as that of the other three (Zhang et al. Citation2006; Lim et al. Citation2008; Yang et al. Citation2008). Furthermore, when using Targetscan, two reports indicated that SALL4 may be a possible target for miR-219-5p (Cheng et al. Citation2015), and miR-107 (He et al. Citation2013). With the aid of gain and loss of function assays, miR-219-5p was shown to inhibit cellular proliferation, invasion and drug resistance by targeting SALL4 expression in colorectal cancers (Cheng et al. Citation2015). On the other hand, miR-107 directly regulated SALL4 expression in glioma cell lines (He et al. Citation2013). And finally, epigenetic regulation of SALL4 has also been reported. Specifically, methylation status of both promoter and the Exon1/intron1 region of the gene, correlates with the expression of SALL4 (Nishino et al. Citation2010; Yang et al. Citation2012; Lin et al. Citation2013; Amabile et al. Citation2015).
SALL4 and CD44 share an interesting relationship with the Wingless/Wnt pathway. In the normal state, SALL4 is directly regulated by the canonical Wnt signalling pathway (Böhm et al. Citation2006). Targeting SALL4 in oesophageal cancers resulted in a diminished Wnt/Beta-catenin pathway with reduced tumorigenicity (He et al. Citation2016). CD44 is also a target of the Wnt pathway (Kwong and Dove Citation2009). In colon cancer, it was shown that the Wnt pathway is the main route for stemness maintenance of cancer cells, usually predicting a poor prognosis (Kwong and Dove Citation2009).
The two proteins also have an interesting relationship with the Epithelial–Mesenchymal transition (EMT) phenomenon in cancer. Evidence shows that a shift from CD44v to CD44s isoforms is essential for the occurrence of EMT process in breast cancer patients (Brown et al. Citation2011). On the other hand, knockdown of the SALL4 and CD44 stemness agents reversed the EMT phenomena; and reduced cellular invasion and metastases in endometrial and hepatocellular cancers, respectively (Mima et al. Citation2013; Liu et al. Citation2015). In gastric cancer, CD44 was identified as a downstream target of SALL4 in cancer cells (Yuan et al. Citation2016). When researchers created a stable knockdown of SALL4, reversal of EMT in gastric cancer cells ensued with decreased expression of stemness genes in those cells and significantly retarded the growth of xenograft tumours in mice (Yuan et al. Citation2016). In their rescue study, restoration of CD44 expression in SALL4 knockdown cells resulted in restoration of the EMT properties of those gastric cells, and greatly accelerated xenograft tumour growth (Yuan et al. Citation2016). In non-small cell lung cancer (NSCLC), SALL4 expression was shown to be positively correlated with CD44 expression in NSCLC, both by immunohistochemistry and RT–PCR (Du et al. Citation2018 Apr Citation25). Those NSCLC cells which expressed CD44, were shown to have a stronger nuclear expression of SALL4; and when NSCLC cells’ SALL4 expression was knocked-down, all stemness-related genes expressions were knocked-down as well, chief among them was CD44 (Du et al. Citation2018 Apr Citation25). Further evidence of the intricate relationship between the two proteins is provided when the CD44 promoter region was cloned and inserted into a luciferase reporter vector (Yuan et al. Citation2016). It was shown that there is region within the CD44 which was critical for SALL4-mediated transactivation of CD44 (Yuan et al. Citation2016).
From the above, the two proteins’ shared involvements in several transformational activities including the EMT phenomenon, the Wnt pathway and the stemness phenomenon; may provide a platform on which a tumour may develop and progress. With co-expression of both proteins in the cytoplasm and nuclei of LSCC cases, this study begs the question if a similar mechanism of action, seen in other tumours, is also at play here for the development and maintenance of laryngeal cancer.
Conclusion
SALL4 is a highly versatile stemness agent, with multiple roles in both health and disease. Its involvement with the stemness phenotype, the EMT phenomenon, and most recently with lncRNAs, makes it a possible target of cancer treatment. This is the first study analysing the expression of SALL4 in laryngeal tumours; along with that of CD44 stemness agent. In line with the other cancers, LSCCs positively express both SALL4 and CD44. This dual positive expression requires further investigation to elucidate the relationship between the two proteins, and with several molecular processes including tumorigenesis, the Wnt pathway; lncRNAs and with the EMT phenomenon in laryngeal cancer; and to further delineate the functions of this biomarker in laryngeal cancer cells using in-vitro as well as in-vivo assays. This may prove to be a new hope to alter the high treatment failures and recurrence rates of laryngeal tumours.
Acknowledgements
The author would like to thank Professor Emad Raddaoui, consultant and senior pathologist at King Khaled University Hospital, King Saud University, Riyadh, Saudi Arabia, for his help in the immunohistochemical scoring of this research work.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
‘The data supporting the findings of this study are available in the figshare repository’ [https://figshare.com/] at https://doi.org/10.6084/m9.figshare.15087936.v1.
References
- Amabile G, et al. 2015. Dissecting the role of aberrant DNA methylation in human leukaemia. Nat Commun. 6:7091. [PubMed: 25997600] Tatetsu et al. Page 11 Gene. Author manuscript; available in PMC 2017 June 15.]
- Baccelli I, Stenzinger A, Vogel V, Pfitzner BM, Klein C, Wallwiener M, Scharpff M, Saini M, Holland–Letz T, Sinn HP, et al. 2014. Co–expression of MET and CD47 is a novel prognosticator for survival of luminal breast cancer patients. Oncotarget. 5:8147–8160.
- Barbato L, Bocchetti M, Di Biase A, Regad T. 2019 Aug 18. Cancer stem cells and targeting strategies cells. 8(8):926. doi:10.3390/cells808092.
- Baumann M, Krause M. 2010. CD44: a cancer stem cell-related biomarker with predictive potential for radiotherapy. Clin Cancer Res. 16(21):5091–5093.
- Böhm J, Sustmann C, Wilhelm C, Kohlhase J. 2006 Sep 29. SALL4 is directly activated by TCF/LEF in the canonical Wnt signaling pathway. Biochem Biophys Res Commun. 348(3):898–907. doi:10.1016/j.bbrc.2006.07.124. Epub 2006 Jul 31. PMID: 16899215.
- Brown R L, Reinke L M, Damerow M S, Perez D, Chodosh L A, Yang J, Cheng C. 2011. CD44 splice isoform switching in human and mouse epithelium is essential for epithelial-mesenchymal transition and breast cancer progression. J Clin Invest. 121:1064–1074.
- Chai L, Liu H, Zhang Z, Wang F, Wang Q, Zhou S, Wang S. 2014. CD44 expression is predictive of poor prognosis in pharyngolaryngeal cancer: systematic review and meta-analysis. Tohoku J Exp Med. 232:9–19.
- Cheng J, Deng R, Zhang P, Wu C, Wu K, Shi L, Liu X, Bai J, Deng M, Shuai X, et al. 2015. miR-219-5p plays a tumor suppressive role in colon cancer by targeting oncogene Sall4. Oncol Rep. 34(4):1923–1932.
- Cheng J, Gao J, Shuai X, Tao K. 2016. Oncogenic protein SALL4 and ZNF217 as prognostic indicators in solid cancers: a meta analysis of individual studies. Oncotarget. 7:24314–24325. doi:10.18632/oncotarget.8237.
- Dawood S, Austin L, Cristofanilli M. 2014 Dec. Cancer stem cells: implications for cancer therapy. Oncology (Williston Park). 28(12):1101–1107, 1110.
- de Jong MC, Pramana J, van der Wal JE, et al. 2010. CD44 expression predicts local recurrence after radiotherapy in larynx cancer. Clin Cancer Res. 16:5329–5338R.
- Di C, Sun J, Zhang H, Zhou P, Kong J. 1 Mar. 2018. High expression of SALL4 is associated with poor prognosis in squamous cell carcinoma of the uterine cervix. Int J Clin Exp Pathol. 11(3):1391–1398.
- Du W, Ni L, Liu B, Wei Y, Lv Y, Qiang S, Dong J, Liu X. 2018 Apr 25. Upregulation of SALL4 by EGFR activation regulates the stemness of CD44-positive lung cancer. Oncogenesis. 7(4):36. doi:10.1038/s41389-018-0045-7. PMID: 29691367; PMCID: PMC5915399.
- Fu Z, Li G, Li Z, Wang Y, Zhao Y, Zheng S, Ye H, Luo Y, Zhao X, Wei L, et al. 2017 May 29. Endogenous miRNA sponge LincRNA-ROR promotes proliferation, invasion and stem cell-like phenotype of pancreatic cancer cells. Cell Death Discov. 3:17004. doi:10.1038/cddiscovery.2017.4. PMID: 28580169; PMCID: PMC5447127.
- Fujii Y, et al. 2012. CDX1 confers intestinal phenotype on gastric epithelial cells via induction of stemnessassociated reprogramming factors SALL4 and KLF5. Proc Natl Acad Sci U S A. 109:20584–20589. PubMed: 23112162].
- Gao L, Yan L, Lin B, Gao J, Liang X, Wang Y, et al. 2011. Enhancive effects of Lewis y antigen on CD44-mediated adhesion and spreading of human ovarian cancer cell line RMG-I. J Exp Clin Cancer Res. 30:15.
- Gao S, Wang P, Hua Y, Xi H, Meng Z, Liu T, Chen Z, Liu L. 2016. ROR functions as a ceRNA to regulate Nanog expression by sponging miR-145 and predicts poor prognosis in pancreatic cancer. Oncotarget. 7:1608–1618. https://www.oncotarget.com/article/6450/text/.
- Greco A, Rizzo M, De Virgilio A, Gallo A, Fusconi M, Pagliuca G, Martellucci S, Turchetta R, De Vincentiis M. 2016. Cancer stem cells in laryngeal cancer: what we know. Eur ArchOto-Rhino-Laryngol. 273:3487–3495.
- Hao L, Yan Zhao Y, Wang Z, et al. 2016. Expression and clinical significance of SALL4 and b-catenin in colorectal cancer. J Mol Hist. 47:117–128. doi:10.1007/s10735-016-9656-5.
- He J, Zhang W, Zhou Q, Zhao T, Song Y. 2013. Chai l and Li Y: Low-expression of microRNA-107 inhibits cell apoptosis in glioma by upregulation of SALL4. Int J Biochem Cell Biol. 45:1962–1973.
- He J, Zhou M, Chen X, Yue D, Yang L, Qin G, Zhang Z, Gao Q, Wang D, Zhang C, et al. 2016 Jun 21. Zhang y inhibition of SALL4 reduces tumorigenicity involving epithelial-mesenchymal transition via Wnt/beta-catenin pathway in esophageal squamous cell carcinoma. J Exp Clin Cancer Res. 35(1):98. doi:10.1186/s13046-016-0378-z.
- Hou P, Zhao Y, Li Z, et al. 2014. LincRNA-ROR induces epithelial-to-mesenchymal transition and contributes to breast cancer tumorigenesis and metastasis. Cell Death Dis. 5:e1287. doi:10.1038/cddis.2014.249.
- Janisiewicz AM, Shin JH, Murillo-Sauca O, Kwok S, Le Q-T, Sunwoo JB. 2012. CD44(+) cells have cancer stem cell-like properties in nasopharyngeal carcinoma. Int Forum Allergy Rhinol. Nov. 2(6):465–470. doi:10.1002/alr.21068. Epub 2012 Aug 7.
- Kokko LL, Hurme S, Maula SM, Alanen K, Grénman R, Kinnunen I, Ventelä S. 2011. Significance of site-specific prognosis of cancer stem cell marker CD44 in head and neck squamous-cell carcinoma. Oral Oncol. 47(6): 510–516.
- Kwong LN, Dove WF. 2009. APC and its modifiers in colon cancer. Adv Exp Med Biol. 14:85–106.
- Li A, et al. 2015. SALL4 is a new target in endometrial cancer. Oncogene. 34:63–72. PubMed: 24336327].
- Lim CY, et al. 2008. Sall4 regulates distinct transcription circuitries in different blastocyst-derived stem cell lineages. Cell Stem Cell. 3:543–554. PubMed: 18804426].
- Lin J, et al. 2013. Aberrant hypomethylation of SALL4 gene in patients with myelodysplastic syndrome. Leuk Res. 37:71–75. PubMed: 23122807].
- Liu L, Zhang J, Yang X, Fang C, Xu H, Xi X. 2015. SALL4 as an epithelial-mesenchymal transition and drug resistance inducer through the regulation of c-Myc in endometrial cancer. PLoS One. 10(9):e0138515. Published 2015 Sep 25. doi:10.1371/journal.pone.0138515.
- Ma Z, Wang YY, Xin HW, Wang L, Arfuso F, Dharmarajan A, Kumar AP, Wang H, Tang FR, Warrier S, et al. 2019 Mar. The expanding roles of long non-coding RNAs in the regulation of cancer stem cells. Int J Biochem Cell Biol. 108:17–20. doi:10.1016/j.biocel.2019.01.003. Epub 2019 Jan 7. PMID: 30630112.
- Masuko K, Okazaki S, Satoh M, Tanaka G, Ikeda T, Torii R, Ueda E, Nakano T, Danbayashi M, Tsuruoka T, et al. 2012. Anti-tumor effect against human cancer xenografts by a fully human monoclonal antibody to a variant 8-epitope of CD44R1 expressed on cancer stem cells. PLoS ONE. 7(1):e29728.
- Mei K, et al. 2009. Diagnostic utility of SALL4 in primary germ cell tumors of the central nervous system: a study of 77 cases. Mod Pathol. 22:1628–1636. PubMed: 19820689].
- Mima K, Hayashi H, Imai K, Kuroki H, Nakagawa S, Okabe H, Chikamoto A, Watanabe M, Beppu T, Baba H. 2013. High CD44s expression is associated with the EMT expression profile and intrahepatic dissemination of hepatocellular carcinoma after local ablation therapy. J Hepatobiliary Pancreat Sci. 20:429–434.
- Nastaly P, et al. 2014. Circulating tumor cells in patients with testicular germ cell tumors. Clin Cancer Res. 20:3830–3841. PubMed: 24634372].
- Nicolè L, Sanavia T, Veronese N, Cappellesso R, Luchini C, Dabrilli P, Fassina A. 2017 Apr 4. Oncofetal gene SALL4 and prognosis in cancer: A systematic review with meta-analysis. Oncotarget. 8(14):22968–22979. doi:10.18632/oncotarget.14952. PMID: 28160555; PMCID: PMC5410278.
- Nie X, Guo E, Wu C. 2019 Apr. SALL4 induces Radioresistance in nasopharyngeal carcinoma via the ATM/Chk2/p53 pathway. Cancer Med. 8(4):1779–1792. doi:10.1002/cam4.2056. Epub 2019 Mar 24.
- Nishino K, et al. 2010. Defining hypo-methylated regions of stem cell-specific promoters in human iPS cells derived from extra-embryonic amnions and lung fibroblasts. PLoS One. 5:e13017. PubMed: 20885964].
- Ouban A, Ahmed A. 2015 Jul. Analysis of the distribution and expression of claudin-1 tight junction protein in the oral cavity. Appl Immunohistochem Mol Morphol. 23(6):444–448. doi:10.1097/PAI.0000000000000104.
- Pan L, Liang W, Gu J, Zang X, Huang Z, Shi H, Chen J, Fu M, Zhang P, Xiao X, et al. 2017 Dec 6. Long noncoding RNA DANCR is activated by SALL4 and promotes the proliferation and invasion of gastric cancer cells. Oncotarget. 9(2):1915–1930. doi:10.18632/oncotarget.23019. PMID: 29416741; PMCID: PMC5788609.
- Qi F, Liu X, Wu H, Yu X, Wei C, Huang X, Ji G, Nie F, Wang K. 2017. Long noncoding AGAP2-AS1 is activated by SP1 and promotes cell proliferation and invasion in gastric cancer. J Hematol Oncol. 10:48.
- Shen H, Li L, Wang D, Yang S, Chen X, Zhou S, Zhong S, Zhao J, Tang J. 2017 Jul 4. Higher expression of SALL4 predicts poor cancer prognosis: a meta-analysis. Cancer Biomark. 19(4):365–373. doi:10.3233/CBM-160052.
- Shen Z, Li Q, Deng H, Lu D, Song H, Guo J. 2014 Sep 22. Long non-coding RNA profiling in laryngeal squamous cell carcinoma and its clinical significance: potential biomarkers for LSCC. PLoS One. 9(9):e108237. doi:10.1371/journal.pone.0108237. eCollection 2014.
- Si X, Zang R, Zhang E, Liu Y, Shi X, Zhang E, Shao L, Li A, Yang N, Han X, et al. 2016. LncRNA H19 confers chemoresistance in ERalpha-positive breast cancer through epigenetic silencing of the pro-apoptotic gene BIK. Oncotarget. 7:81452–81462. doi:10.18632/oncotarget.13263.
- Tatetsu H, Kong NR, Chong G, et al. 2016 Jun 15. SALL4, the missing link between stem cells, development and cancer. J. 584(2):111–119. doi:10.1016/j.gene.2016.02.019. Epub 2016 Feb 16.
- Ueno S, et al. 2014. Aberrant expression of SALL4 in acute B cell lymphoblastic leukemia: mechanism, function, and implication for a potential novel therapeutic target. Exp Hematol. 42:307–316. e308. [PubMed: 24463278].
- Wang F, et al. 2013. Stem cell factor SALL4, a potential prognostic marker for myelodysplastic syndromes. J Hematol Oncol. 6:73. PubMed: 24283704].
- Wu H, Liu C, Fan X, et al. 2017 Aug 23. Spalt-like Transcription Factor 4 as a potential diagnostic and prognostic marker of colorectal cancer. Cancer Biomark. 20(2):191–198. doi:10.3233/CBM-170204.
- Yang J, et al. 2008. Genome-wide analysis reveals Sall4 to be a major regulator of pluripotency in murineembryonic stem cells. Proc Natl Acad Sci U S A. 105:19756–19761. PubMed: 19060217].
- Yang J, Corsello TR, Ma Y. 2012. Stem cell gene SALL4 suppresses transcription through recruitment of DNA methyltransferases. J Biol Chem. 287:1996–2005. PubMed: 22128185].
- Yong KJ, Gao C, Lim JS, Yan B, Yang H, Dimitrov T, Kawasaki A, Ong CW, Wong KF, Lee S, et al. 2013 Jun 13. Oncofetal gene SALL4 in aggressive hepatocellular carcinoma. N Engl J Med. 368(24):2266–2276. doi:10.1056/NEJMoa1300297.
- Yu D, Jin C, Liu Y, Yang J, Zhao Y, Wang H, Zhao X, Cheng J, Liu X, Liu C. 2013 Dec. Tumour clinical implications of cancer stem cell-like side population cells in human laryngeal cancer. Biol. 34(6):3603–3610. doi:10.1007/s13277-013-0941-6. Epub 2013 Jun 27.
- Yu D, Jin C, Liu Y, Yang J, Zhao Y, Wang H, Zhao X, Cheng J, Liu X, Liu C. 2013. Clinical implications of cancer stem cell-like side population cells in human laryngeal cancer. Tumour Biol. 34(6):3603–3610.
- Yuan JH, Li WX, Hu C, Zhang B. 2020 Jul. Upregulation of SNHG12 accelerates cell proliferation, migration, invasion and restrain cell apoptosis in breast cancer by enhancing regulating SALL4 expression via sponging miR-15a-5p. Neoplasma. 67(4):861–870. doi:10.4149/neo_2020_190808N731. Epub 2020 May 6. PMID: 32386479.
- Yuan X, Zhang X X, Zhang W, Liang W, Zhang P, Shi H, Zhang B, Shao M, Yan Y, Qian H, Xu W. 2016. SALL4 promotes gastric cancer progression through activating CD44 expression. Oncogenesis. 5:e268. doi:10.1038/oncsis.2016.69.
- Zhang J, et al. 2006. Sall4 modulates embryonic stem cell pluripotency and early embryonic development by the transcriptional regulation of Pou5f1. Nat Cell Biol. 8:1114–1123. PubMed: 16980957].
- Zhang L, et al. 2014. SALL4, a novel marker for human gastric carcinogenesis and metastasis. Oncogene. 33:5491–5500. PubMed: 24276240].
- Zhang X, Yuan X, Zhu W, Qian H, Xu W. 2015. SALL4: an emerging cancer biomarker and target. Cancer Lett. 357:55–62.