Abstract
Captive breeding is among the most effective approaches for ensuring the conservation of numerous endangered species. However, only a few studies have examined the effects of captive maintenance of animals at the molecular level. In this study, we adopted the isobaric tags for relative and absolute quantification (iTRAQ) approach to compare protein expression in the blood of endangered spotted seal pups (Phoca largha) inhabiting different environments. We identified 519 proteins from 2,628 peptides, the expression of 158 of which differed significantly between short-term captive and wild seals. In addition, 140 proteins were identified as being differentially expressed between short-term and long-term captive seals, whereas 235 proteins were differentially expressed between the wild and long-term captive groups (p < 0.05). In wild seal pups, proteins associated with the maintenance of cardiomyocyte cell stability and integrity, protection of the heart, and prevention of anemia were upregulated, reflecting the high stress and energetic costs associated with movement and foraging. In addition, significant differences were detected among the three groups with respect to immune-related proteins. The results of this study advance our current understanding of protein expression in spotted seals and provide data that will contribute to the future conservation of this species.
Introduction
Spotted seals (Phoca largha) are the only pinniped species that reproduces in China. They are mainly distributed in the Bohai Sea and the northern part of the Yellow Sea, although can occasionally be found in the East and South China seas. Breeding takes place in winter on ice flows in Liaodong Bay along the Bohai Sea coast Wang et al. (Citation2008). However, as a consequence of habitat destruction, loss of breeding sites on the ice, and illegal capture, the numbers of spotted seal have declined in recent times. It is estimated that there were approximately 8,000 spotted seals in the Yellow Sea during the early 1940s; however, by the 1980s, numbers had declined to 2,300 Xiang et al. (Citation2010). Similarly, the numbers of spotted seals in Liaodong Bay have undergone a marked decline over the past few decades (Gao et al. (Citation2015), and the species is currently classified as critically endangered in China and South Korea, in addition to being designated as a national first-level key protected species in China Wang et al. (Citation2008); Gao et al. (Citation2015); Gao et al. (Citation2013).
Given the current threats posed to natural populations worldwide, captive breeding is seen as one of the most effective approaches to ensure the conservation of numerous endangered species, and is a tool that has been increasingly adopted in China over the past few decades. For example, eight spotted seal reserves have been established in coastal areas, such as Dalian, Liaoning Province; Panjin, Liaoning Province; and Miaodao, Shandong Province in China Wang et al. (Citation2008), and since 2012, the number of spotted seals raised in captivity in China has increased from 400 to more than 1,000, accounting for approximately 50% of the entire population in the country Trukhin (Citation2003); Aerts et al. (Citation2013); Yang et al. (Citation2017).
However, although this type of artificial intervention has undeniably ensured the survival of spotted seals, there is extensive experimental evidence to indicate that different living environments can affect the physiological and biochemical characteristics of a diverse range of animals. For example, captive Sunda pangolins (Manis javanica) have been found to have significantly lower white blood cell counts, neutrophil counts, and alanine phosphatase and phosphorus levels; significantly higher packed cell volumes; and elevated levels of total protein, globulin, and blood urea nitrogen than rescued wild Sunda pangolins Ahmad et al. (Citation2020). Moreover, three groups of harbor seals (Phoca vitulina; free-ranging animals, rehabilitated pups, and captive seals in the Friedrichskoog Seal Center in Germany) showed significantly different hematological profiles Hasselmeier et al. (Citation2008). In addition, individuals of the same species can often display different metabolic levels and high phenotypic variations in response to varying environmental conditions. Similarly, compared with conditions in their natural habitats, captive environments are associated with reduced standard metabolic rates, maximum metabolic rates, and absolute aerobic scope, and altered morphological traits in the endangered Spanish toothcarp (Aphanius iberus) Latorre et al. (Citation2020).
In this context, extensive molecular studies have been conducted on seals. For example, transcriptomic studies have been undertaken to evaluate the brain tissues of seven newborn harbor seals that had died from unknown causes to determine the cause of death Rosales and Thurber (Citation2016), and to analyze the blood, muscle tissue, and adipose tissue of the northern and southern elephant seals (Mirounga leonina and Mirounga angustirostris, respectively) Khudyakov et al. (Citation2015); Martinez et al. (Citation2018); Kim et al. (Citation2019). In addition, transcriptome analysis has been used to identify key metabolic changes in the brains of hooded seals (Cystophora cristata) in response to hypoxia and reoxygenation Hoff et al. (Citation2017). Contrastingly, investigations on spotted seals have tended to focus on populations and distribution in the field, genetic analysis, and habitat evaluation Wang et al. (Citation2008); Han et al. (Citation2010), whereas relatively few molecular studies have been conducted, particularly proteomic-based investigations. Studies at the protein level can provide information regarding the presence of different protein isoforms or the post-translational modifications of proteins, and given that proteins have identifiable biological functions, determining protein expression profiles represents a powerful tool facilitating species characterization. However, to date, few studies have examined the differences in physiological and biochemical characteristics of spotted seals using molecular approaches.
To the best of our knowledge, the present study is the first in which a quantitative proteomic profiling using isobaric tags for relative and absolute quantification (iTRAQ) approach has been adopted to analyze the blood of spotted seal (P. largha) pups inhabiting different environments. The results obtained provide an overview of differences in the whole blood proteomes of P. largha seals living under different environmental conditions. Moreover, we also identified key functional proteins, the expression profiles of which might differ in these different environments. We believe that this information will advance our current understanding of protein expression profiles in spotted seals and provide data that could potentially contribute to the effective conservation of this species.
Materials and methods
Animals
During a Spotted Seal Rescue Survey conducted in 2019, blood samples were obtained from wild P. largha pups inhabiting Liaodong Bay, China. Given their white lanugo and the fact that they were independent of their mothers, it is assumed that although younger than 1 month of age, these individuals were no longer being nursed. The captive P. largha pups used in the present study had been born in captivity and were being maintained at the Dalian Sun Asia Aquarium, Liaoning Province, China. As a third sample population, wild P. largha pups were captured and temporarily maintained in an aquarium for 2 months. All seals were fed capelin (Mallotus villosus) and Atlantic herring (Clupea harengus) (m:m) at a 1:1 ratio, with the daily amount of feed being adjusted to approximately 8% of the pups’ weight. The ontogenetic differences among P. largha pups in the three populations are presented in Table .
Table 1. Ontogenetic differences in Phoca largha pups in three environments.
Analysis of blood, an important component of the immune system, is a useful approach for genetic studies. Approximately 3 mL of blood was collected from the hind flipper veins of animals restrained on a V-shaped bench and caused little distress to the sampled animals. The same handling protocol was used for P. largha pups in all three experimental groups, for each of which, we performed three biological replicate analyses. Blood samples (5 mL) were placed separately in RNase-free tubes containing silicone separation gel (Kang Wei Si Medical Instrument CO., LTD, Hebei, China). After centrifugation at 12,000 × g for 5 min at 4°C, the serum was separated and placed in cryogenic lyophilization tubes that were immediately frozen in liquid nitrogen for storage until protein extraction. All operations were performed in accordance with the Chinese national standard Rearing specifications for spotted seals (SC/T 9606-2018).
Protein digestion and iTRAQ labeling
Initially, the three biological replicate samples for each experimental group were respectively mixed to obtain total samples for each of the three populations. The most abundant serum proteins were extracted using a ProteoExtract™ Albumin/IgG Removal Kit (Merck KGaA, Darmstadt, Germany). For each sample, 30 μL of serum was diluted with 600 µL of buffer and loaded onto an affinity column. The eluate thus obtained was collected in 10-kDa ultrafiltration tubes for concentration, and protein quantification was performed using the BCA method. Ten micrograms of isolated protein from each sample was used for SDS-PAGE.
A further 200 µg of proteins from each sample was used for trypsin digestion using the filter-aided proteome preparation method Wiśniewski et al. (Citation2009). The peptic digests thus obtained were subsequently desalted using a C18 Cartridge (Sigma-Aldrich, Saint Louis, USA), and following lyophilization, 40 μL of dissolution buffer was added to redissolve the peptides, which were quantified spectrophotometrically at wavelength of 280 nm.
iTRAQ labeling was performed using an iTRAQ Reagent-8PLEX Multiplex Kit according to the manufacturer’s instructions (Applied Biosystems, Foster City, USA). The proteins in the wild, short-term captive, and long-term captive P. largha pup groups were labeled with reagents 115, 114, and 116, respectively.
Strong cation exchange fractionation and liquid chromatography–tandem mass spectrometry analysis
The iTRAQ-labeled samples were fractionated based on strong cation exchange chromatography using a polysulphoethyl column (4.6 mm × 100 mm, 5 μm, 200 Å; PolyLC Inc., Columbia, MD, USA) in an AKTA Purifier 100 (GE Healthcare, Piscataway, USA), the flow rate in which was set to 1 mL/min. The column was equilibrated with buffer A (10 mM KH2PO4, 25% ACN, pH 3.0), and samples were eluted with a linear gradient of 0% to 10% buffer B (10 mM KH2PO4, 500 mM KCl, 25% ACN, pH 3.0) for 25 min, and then a linear gradient of 10% to 45% buffer B for 17 min, followed by a 10 min linear gradient to 100%, at which the samples were held for 8 min. During the elution process, absorbance was monitored at 214 nm, with eluates being collected at 1-min intervals. Following lyophilization, a C18 Cartridge (Sigma-Aldrich, Saint Louis, USA) was used for desalination.
Fractions were separated using a nano HPLC EASY-nLC system (Thermo Finnigan, Hemel Hempstead, UK) incorporating two Thermo Fisher Scientific (Hemel Hempstead, UK) EASY columns (2 cm × 100, 5 μm-C18 for sampling and 75 μm × 100 mm, 3 μm-C18 for analysis). Flow rates were set to 250 nL/min, and the columns were equilibrated with 95% mobile phase A (0.1% formic acid in water). The fractions were eluted in a linear gradient starting from 0% to 35% mobile phase B (0.1% formic acid in 84% acetonitrile) for 50 min, followed by an 8 min linear gradient to 100%, at which the samples were held for 2 min. Mass spectrometry (MS) analysis of eluted samples (peptides) was carried out using a Q-Exactive mass spectrometer (Thermo Finnigan). Full MS survey scans were performed for a mass range of 300–1,800 m/z with a resolution of 70,000 at m/z 200, for which the automatic gain control target was 3e6, Maximum injection time was set to 10 ms, and dynamic exclusion was set to 40 s. Having performed each full survey scan, we collected the 10 most intense signals. Higher-energy collisional dissociation fragmentation was used for tandem mass spectrometry (MS/MS) at a resolution of 17,500 and 200 m/z, and the normalized collision energy was set to 30 eV with a 0.1% underfill ratio.
Data analysis
The raw files were analyzed and proteins identified using Proteome Discoverer v.1.4 software (Thermo Fisher Scientific, Karlsruhe, BW, Germany) and the MASCOT v.2.2 search engine (Matrix Science Ltd, London, UK). The protein identification and quantitation parameters are shown in Table . Hierarchical cluster analysis was used to assess the differentially expressed proteins (DEPs) for significant down- or upregulation relative to their levels in the comparison group, and the data are presented in the form of Heatmaps. The screening criteria for DEPs was a fold change > 1.5 or fold change < 0.667 (p < 0.05). For downstream analysis, the proteins were annotated using the following databases: Gene Ontology (GO), Kyoto Encyclopedia of Genes and Genomes (KEGG), and protein–protein interaction (PPI) networks.
Table 2. Protein identification and quantitation parameters.
Ethics approval
The protocols for sample collection from spotted seals were approved by the Ministry of Agriculture and Rural Affairs of the People’s Republic of China (permit number: 1376).
Results
Protein profiling
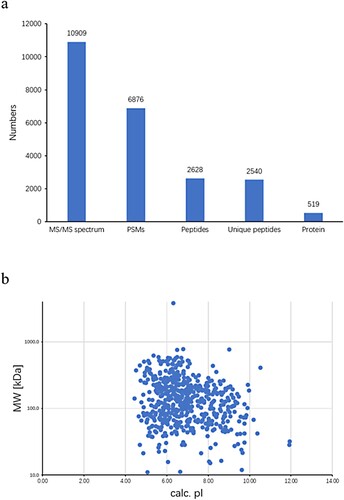
For the purposes of protein identification, we used Phocidae as the protein database taxon. iTRAQ analyses revealed 6,876 peptide spectrum-matches for the total 10,909 spectra. Among the matches, 519 unique proteins were identified among 2,628 peptides (Figure a), among which, there were 63 proteins with a molecular mass of between 10 and 50 kDa, 137 between 50 and 100 kDa, 93 between 100 and 150 kDa, 77 between 150 and 200 kDa, and 149 with a mass greater than 200 kDa (Figure b).
iTRAQ analysis and identification of DEPs
We used a 1.5-fold increase or 0.667-fold decrease in protein expression as criteria for defining a physiologically significant change. In total, 158 DEPs (p < 0.05) were identified between the short-term captive and wild groups, among which 86 and 72 were up- and downregulated, respectively (Figure S1 a); 140 DEPs (p < 0.05) were identified between the short-term captive and long-term captive groups, among which 62 and 78 were up- and downregulated, respectively (Figure S1 b); and 235 DEPs (p < 0.05) were identified between the wild and long-term captive groups, among which 117 and 118 were up- and downregulated, respectively (Figure S1 c).
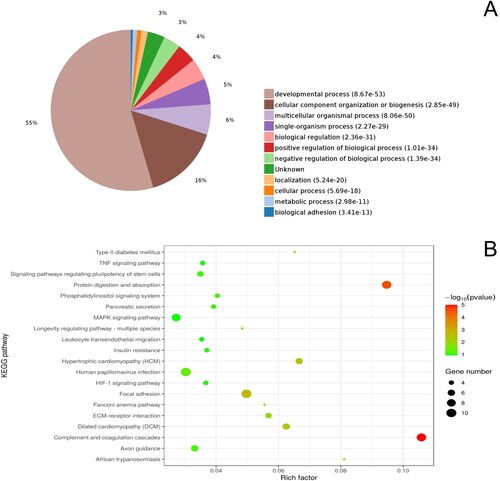
GO enrichment analysis performed for the short-term captive and wild groups revealed that 3,693, 555, and 586 proteins were enriched in the three categories biological processes, cell components, and molecular functions, respectively. Of these, the differential expression of 2,144, 351, and 256 proteins was statistically significant (Table S1). Among biological processes, the top five categories with the highest number of enriched DEPs were ‘multicellular organismal process’ (61%), ‘cellular component organization or biogenesis’ (17%), ‘biological regulation’ (8%), ‘single-organism process’ (4%), and ‘positive regulation of biological process’ (3%) (Figure A). Moreover, we identified 181 KEGG pathways that were enriched in this dataset, of which 13 were statistically significant (Table S1). Pathways most highly enriched in DEPs were focal adhesion (9), MAPK signaling pathway (7), tight junction (5), protein digestion and absorption (5), and ECM-receptor interaction (4) (Figure B). PPI network analysis of the identified DEPs was performed using STRING software, the results of which are shown in Table S1. The search results indicated that PIK3CD had the highest number of predicted interactions with KEGG pathways, having interactive associations with six pathways. Among other proteins, COL6A3, COL9A3, and COL6A6 were each established to interact with four pathways Figures , .
Figure 2. GO functional enrichment analysis (A) and KEGG pathway enrichment analysis (B) of differentially expressed proteins between the short-term captive and wild groups.
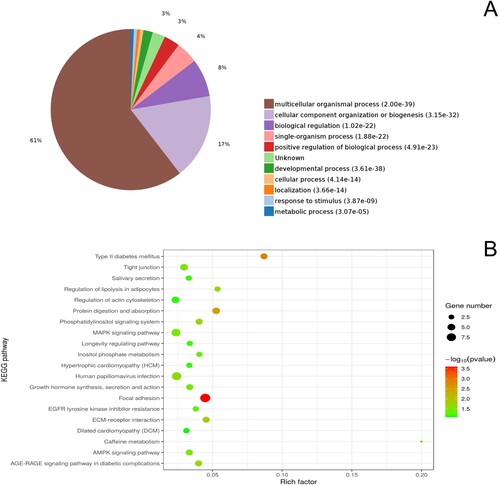
Figure 3. GO functional enrichment analysis (A) and KEGG pathway enrichment analysis (B) of differentially expressed proteins between the short-term captive and long-term captive groups.
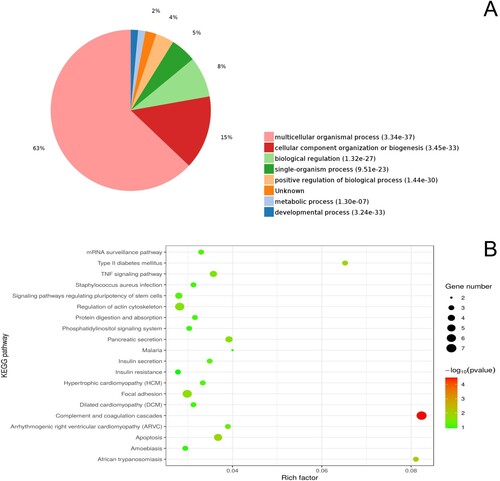
Figure 4. GO functional enrichment analysis (A) and KEGG pathway enrichment analysis (B) of differentially expressed proteins between the wild and long-term captive groups.
For the short-term captive and long-term captive group comparison, GO enrichment analysis indicated that 3,905, 530, and 599 proteins were enriched in the categories biological processes, cell components, and molecular functions, respectively, of which, 2,579, 356, and 275 were statistically significant, respectively (Table S1). The five biological process categories with the highest number of enriched DEPs were ‘multicellular organismal process’ (63%), ‘cellular component organization or biogenesis’ (15%), ‘biological regulation’ (8%), ‘single-organism process’ (5%), and ‘positive regulation of biological process’ (4%). Moreover, we identified 183 enriched KEGG pathways, of which eight were statistically significant (Table S1). KEGG analysis revealed that the pathways most highly enriched in DEPs were complement and coagulation cascades (7), regulation of actin cytoskeleton (6), focal adhesion (6), apoptosis (5), and TNF signaling pathway (4). PPI network analysis of the identified DEPs revealed that PIK3CD interacts with five KEGG pathways, and RYR2 and KMG1 each interact with three pathways (Figure S2).
With respect to the wild and long-term captive groups, GO enrichment analysis indicated that 4,534, 642, and 734 proteins were enriched for the categories biological processes, cell components, and molecular functions, respectively, and of these, 2,841, 412, and 333 were statistically significant, respectively (Table S1). Among biological processes, the five categories most highly enriched in DEPs were ‘developmental process’ (55%), ‘cellular component organization or biogenesis’ (16%), ‘multicellular organismal process’ (6%), ‘single-organism process’ (5%), and ‘biological regulation’ (4%). KEGG pathway analysis revealed 203 pathways to be DEP-enriched, of which nine were statistically significant (Table S1). The KEGG pathways most highly enriched in DEPs were focal adhesion (10), complement and coagulation cascades (9), protein digestion and absorption (9), and ECM-receptor interaction (5) (Figure S2). PPI networks of the identified DEPs indicated ITGA5, COL9A3, and COL6A6 to be most interactive, each of which was associated with four KEGG pathways (Figure S2).
DEPs
We found that the expression of proteins associated with cytoskeletal function differed significantly among the wild, short-term captive, and long-term captive groups. Among these, phosphatidylinositol 3-kinase, ankyrin, and six types of collagen alpha were significantly upregulated, whereas filamin-C and spectrin alpha chain were significantly downregulated (Table ). Moreover, the expression of immune-related proteins, including carboxypeptidase B2, kininogen-1, three zinc finger proteins, and two RING finger proteins, was found to be lower in the long-term captive group than in the wild group, whereas in contrast, the expression of fibrinogen alpha chain, E3 ubiquitin-protein ligase, and plasma kallikrein was the highest in the long-term captive group and the lowest in the wild group. Related proteins, which are enriched during growth, including neurolysin, keratin, and clusterin, were significantly upregulated in the wild group, whereas the levels of nebulin, vasohibin, myosin-9, and fibrillin were downregulated in this group. In addition, the levels of insulin receptor substrate 4 expression were observed to be the highest in the wild group, being 4.3-fold and 11.61-fold higher than those in the long-term and short-term captive groups, respectively (Table S2).
Table 3. The screening criteria for DEP (fold change > 3.0 or fold change < 0.5) from wild group /captive group(WG/CG), captive group /temporary-captive group (WG/TCG) and captive group /temporary captive group (WG/TCG).
Discussion
Our GO enrichment analyses revealed that proteins characterized by differential expression between the wild and the long-term captive groups were most enriched in the category ‘developmental process,’ accounting for 55% of all identified DEPs. It can be speculated that such differences reflect the contrasting environments of these seals, given that natural populations are generally more exposed to potential stressors that affect the growth and development of animals Smith and Boesch (Citation2011). In addition, wild animals tend to incur higher energy costs associated with movement and foraging, and consequently have relatively less energy available to support bodily growth Zihlman et al. (Citation2007). Further, it is noteworthy that keratin, neurolysin, and clusterin are among the upregulated DEPs identified as being enriched in the category ‘developmental process’ in the wild group. Keratin is one of the most abundant proteins in epithelial cells and is the main protein comprising hair, horns, claws, and skin. Thus, it is plausible that organisms might maintain large quantities of buffer keratin to cope with the physical, chemical, and microbial pressures imposed by the external environment Toivola et al. (Citation2010); Pan et al. (Citation2013). Pups in the wild group were less than 1 month old and had retained a white lanugo, which serves as a protective camouflage against the background of snow and ice in their native nursery environment. Neurolysin is associated with neuronal cytoplasm and membranes Fontenele-Neto et al. (Citation2001); Woulfe et al. (Citation1992) and also contributes to central and peripheral neurotensin-mediated in vivo functions Jirácek et al. (Citation1997) It has been established that large amounts of neurotensin in the brain can protect animals against strokes Jayaraman et al. (Citation2020). Clusterin is also expressed in the brain; its expression has been shown to be induced in response to stress or inflammation and has been identified as the second most abundantly expressed apolipoprotein putatively involved in the pathology of late-onset Alzheimer’s disease (LOAD) Woody and Zhao (Citation2016). It is suggested that this protein might modulate LOAD risk via regulation of cholesterol metabolism, and that it is also involved in key LOAD-related processes, including Aβ regulation, inflammation, and apoptosis Moon et al. (Citation2021). Consequently, given the more pronounced environmental pressures to which seal pups are exposed in their natural habitats, it is conceivable that enhanced upregulation of these related proteins in wild seals contributes to the avoidance or tolerance of environmental stress.
KEGG pathway analysis revealed that in all three seal groups, the focal adhesion and type II diabetes mellitus pathways were notably enriched in DEPs. The focal adhesion pathway is an important signaling pathway that plays pivotal roles in cell proliferation, differentiation, motility, migration, and survival Block et al. (Citation2009); Cebrian-Serrano et al. (Citation2014); Saeed-Zidane et al. (Citation2019). Among the intermediaries of this pathway, we identified that cytoskeleton-related DEPs, such as filamin-C, calpain-2, and spectrin, exhibited the highest expression in wild group seals. Filamin-C is a muscle-specific filamin, a class of actin-binding proteins that function by interacting with transmembrane proteins Thompson et al. (Citation2000). It is specifically expressed in cardiomyocytes and skeletal myocytes and has been established to be involved in the maintenance of structural integrity Shao et al. (Citation2016). A lack of filamin-C results in severe defects in myogenesis and myotube structure Dalkilic et al. (Citation2006). Calpains belong to a family of calcium-dependent cysteine proteases with limited proteolytic activity Carragher (Citation2006). Calpain activity is required for the limited proteolysis of several substrates, including paxillin, FAK, talin, cortactin, and spectrin, which facilitates cytoskeletal remodeling, focal adhesion disassembly, and membrane protrusion Chan et al. (Citation2010). Calpain-2 has been suggested to induce death and survival signaling Ho et al. (Citation2012); Franco et al. (Citation2004), and has been reported to protect the heart against hemodynamic stress in a mouse model of pressure overload-induced myocardial dysfunction. Similar cardioprotective effects associated with upregulated calpain-2 have been observed in response to heat stress–induced injury and doxorubicin-induced cardiac injury Liu et al. (Citation2019); Zheng et al. (Citation2019). Spectrin is a cytoskeletal protein that is best characterized in mammalian erythrocytes and neuronal cells Goodman et al. (Citation1988), with the findings of previous studies indicating that this protein is a multi-faceted molecule with a host of functions. For example, spectrin has been demonstrated to be involved in cytoskeletal development and the maintenance of cell stability Patel-Hett et al. (Citation2011), and is identified as a major functional component responsible for the maintenance of erythrocyte cell membrane elasticity and strength Patra et al. (Citation2015). Moreover, it can bind to hemoglobin and other heme-containing proteins Datta et al. (Citation2003). The significant upregulation of these proteins in wild spotted seal pups is thus assumed to reflect their important role in maintaining the stability and integrity of cardiomyocytes, the prevention of anemia, and protection of the heart in turbulent natural environments.
With respect to the type II diabetes mellitus pathway, we identified insulin receptor substrate 4 (IRS-4) and phosphatidylinositol 3-kinase (PI3K) as being the common DEPs among all three seal groups, with the expression of IRS-4 being the highest in the wild group. Insulin receptor substrates play important roles in maintaining cell growth, division, and metabolism and are key intermediaries of insulin receptor-mediated signaling. It is assumed that proteins that interacts with these receptors, such as PIK3, are implicated in insulin signaling Sano et al. (Citation2002). IRS-4 would appear to play an important role in cell proliferation, given that it is substantially upregulated during rat liver regeneration after partial hepatectomy Tsuruzoe et al. (Citation2001). Previous studies have also revealed that enhanced expression of IRS-3 and IRS-4 might contribute to insulin resistance, leading to diabetes. In particular, upregulation of IRS-4 leads to impaired insulin signaling, the reduced sensitivity of pituitary cells to insulin, and abnormal insulin secretion Tsuruzoe et al. (Citation2001). However, this putative activity is in need of further confirmation, particularly in light of the findings that IRS-4 knockout mice have only mild growth and reproductive defects, thereby indicating a limited role for IRS-4 in glucose homeostasis Sadagurski et al. (Citation2014). We found expression of this protein to be the highest in wild spotted seal pups, which we suspect could be attributable to the rapid growth of pups at 1 month of age, within an environment in which they are potentially exposed to multiple diseases that could impair growth and survival.
Interestingly, in our comparative analyses of protein expression in the wild and short-term captive seals with that in long-term captive seals, we detected the enrichment of certain DEPs in the complement and coagulation cascade pathways. Among these, immune-related proteins, carboxypeptidase B2, kininogen-1, zinc finger protein, and RING finger protein were found to be expressed at higher levels in the wild group seals than in either of the two captive groups. In this respect, it has previously been demonstrated that maternal malnutrition disrupts the complement and coagulation cascade pathways Chen et al. (Citation2019). There is also circumstantial evidence to indicate that consuming human milk promotes secretory IgA synthesis in the mucosal membranes of newborns. Furthermore, numerous immunosuppressive factors have been identified in maternal colostrum, which might protect the newborn immune system against overstimulation by multiple environmental antigens. In addition, the transmission of antibodies from mother to newborns via milk has been established to confer passive immunity Saha et al. (Citation1994).
Similar to humans, seals gain a certain degree of immunity from their mothers and retain this even after the cessation of nursing. Compared with their native counterparts, seals maintained in captivity over the long term are exposed to relatively stable, and presumably less stressful, environments, which could well account for the observed differential expression of immune-related proteins among the three groups. However, one of the drawbacks of maintenance in captivity is the potentially heightened risk of disease, owing to the close proximity of other animals. The expression of fibrinogen alpha chain (FGA) in spotted seal pups in the long-term captive group was found to be significantly higher than that in the other two groups, and it has been established that high expression of FGA can cause endometriosis Chen et al. (Citation2019). Fibrinogen is also a potential serum protein biomarker for depressive disorder Kang et al. (Citation2019). Moreover, studies have shown that an upregulated expression of serum FGA is involved in pathological processes associated with osteoarthritis, rheumatoid arthritis, gastric cancer, and other diseases Olumuyiwa et al. (Citation2019). The findings of a proteomic study have also indicated that FGA is highly expressed in the background of steroid-induced necrosis of the femoral head Liu et al. (Citation2020). Consequently, conservation efforts focusing on the rescue of wild seals and raising seals in captivity should ideally take into consideration both their physical and mental health, and in this respect, the immunity of captive-reared spotted seal pups could be suitably enhanced through the provision of relevant food additives or other methods.
In conclusion, the present study is the first in which the iTRAQ method has been performed for quantitative proteomic profiling of serum samples from spotted seal (P. largha) pups inhabiting different environments. Significant differences were detected among seals living in the wild and in captivity with respect to a range of proteins, including those associated with growth, the cytoskeleton, and immune function, The results obtained in this study will advance our current understanding of the protein expression profile of spotted seals, provide data that will contribute to the protection and conservation of this species, and lay the foundations for future research.
Supplemental Material
Download Zip (2.7 MB)Acknowledgments
We thank Panjin Guanghe Fisheries Co., Ltd and Dalian Sun Asia Aquarium for their assistance in collecting spotted seal blood samples.
Author contributions
Jiashen Tian, Yingdong Li, and Zhichuang Lu designed the study; Xiang Li, Xin Li, Zhen Wang and Zhongren Kong performed the experiments; Xiang Li, Shengjiu Zhang and Xin Li analyzed the data; Jiashen Tian and Yingdong Li wrote the paper.
Data availability statement
The datasets supporting the conclusions of this study are included within the article and the additional files. The mass spectrometry proteomic data have been deposited with the ProteomeXchange Consortium [http://www.proteomexchange.org/] via the PRIDE partner repository (dataset identifier PXD031310). All analyzed data are available from the corresponding author upon reasonable request.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
References
- Aerts LAM, McFarland AE, Watts BH, Lomac-MacNair KS, Seiser PE, Wisdom SS, Kirk AV, Schudel CA. 2013. Marine mammal distribution and abundance in an offshore sub-region of the northeastern chukchi Sea during the open-water season. Cont Shelf Res. 67:116–126.
- Ahmad A, Sekar S, Oh P, Samsuddin S. 2020. Hematology and serum biochemistry of captive Sunda pangolin (manis javanica) in wildlife reserves Singapore. J Vet Med Sci. 83(2):309–314.
- Block J, Bonilla L, Hansen PJ. 2009. Effect of addition of hyaluronan to embryo culture medium on survival of bovine embryos in vitro following vitrification and establishment of pregnancy after transfer to recipients. Theriogenology. 71:1063–1071.
- Carragher N. 2006. Calpain inhibition: A therapeutic strategy targeting multiple disease states. Curr Pharm Des. 12:615–638.
- Cebrian-Serrano A, Salvador I, Silvestre MA. 2014. Beneficial effect of Two culture systems with small groups of embryos on the development and quality of In vitro-produced bovine embryos. Anat Histol Embryol. 43:22–30.
- Chan K, Bennin D, Huttenlocher A. 2010. Regulation of adhesion dynamics by calpain-mediated proteolysis of focal adhesion kinase (FAK). J Biol Chem. 285:11418–11426.
- Chen Y, Li H, Cheng H-y, Rui-qiong M, Ye X, Cui H, Hong-lan Z, Chang X-h. 2019. Fibrinogen alpha chain is up-regulated and affects the pathogenesis of endometriosis. Reprod Biomed Online. 39:893–904.
- Dalkilic I, Schienda J, Thompson T, Kunkel L. 2006. Loss of filaminC (FLNc) results in severe defects in myogenesis and myotube structure. Mol Cell Biol. 26:6522–6534.
- Datta P, Chakrabarty SB, Chakrabarty A, Chakrabarti A. 2003. Interaction of erythroid spectrin with hemoglobin variants: implications in β-thalassemia. Blood Cells, Molecules, and Diseases. 30:248–253.
- Fontenele-Neto JD, Massarelli E, Garrido P, Beaudet A, Ferro E. 2001. Comparative fine structural distribution of endopeptidase 24.15 (EC3.4.24.15) and 24.16 (EC3.4.24.16) in rat brain. J Comp Neurol. 438:399–410.
- Franco S, Perrin B, Huttenlocher A. 2004. Isoform specific function of calpain 2 in regulating membrane protrusion. Exp Cell Res. 299:179–187.
- Gao X, Han J, Lu Z, Li Y, He C. 2013. De novo assembly and characterization of spotted seal Phoca largha transcriptome using illumina paired-end sequencing. Comp Biochem Phys D. 8:103–110.
- Gao XG, Han JB, Lu ZC, Zhang PJ, He CB. 2015. Short communication sequence variation and gene duplication at the MHC DRB loci of the spotted seal Phoca largha. Genet Mol Res. 14:2055–2062.
- Goodman SR, Krebs KE, Whitfield CF, Riederer BM, Zagon IS, Kay MMB. 1988. Spectrin and related molecule. Critical Reviews in Biochemistry. 23:171–234.
- Han J-B, Sun F-Y, Gao X-G, He C-B, Wang P-L, Ma Z-Q, Zhao HW, Wang C, Ma P, Wang Z. 2010. Low microsatellite variation in spotted seal (Phoca largha) shows a decrease in population size in the Liaodong gulf colony. Ann Zool Fenn. 47(1):15–27.
- Hasselmeier I, Fonfara S, Driver J, Siebert U. 2008. Differential hematology profiles of free-ranging, rehabilitated, and captive harbor seals (<I>Phoca vitulina</I>) of the German north Sea. Aquat Mamm. 34:149–156.
- Ho W-c, Pikor L, Gao Y, Elliott BE, Greer PA. 2012. Calpain 2 regulates akt-foxo-p27Kip1 protein signaling pathway in mammary carcinoma. J Biol Chem. 287:15458–15465.
- Hoff MLM, Fabrizius A, Czech-Damal NU, Folkow LP, Burmester T. 2017. Transcriptome analysis identifies key metabolic changes in the hooded seal (cystophora cristata) brain in response to hypoxia and reoxygenation. PLoS One. 12(1):e0169366.
- Jayaraman S, Al Shoyaib A, Kocot J, Villalba H, Alamri FF, Rashid M, Wangler NJ, Chowdhury EA, German N, Arumugam TV, et al. 2020. Peptidase neurolysin functions to preserve the brain after ischemic stroke in male mice. J Neurochem. 153:120–137.
- Jirácek J, Noble F, Loog M, Roques B-P, Dive V, Vincent JP, Checler F. 1997. Effect of a novel selective and potent phosphinic peptide inhibitor of endopeptidase 3.4.24.16 on neurotensin-induced analgesia and neuronal inactivation. Br J Pharmacol. 121:705–710.
- Kang G, Zhang Y, Liu R, Li R, Kang Q, Zhu X, Yan L, Yu Y, Yu Q. 2019. Fibrinogen and kininogen are potential serum protein biomarkers for depressive disorder. Clin Lab. 65(10):e190312.
- Khudyakov JI, Preeyanon L, Champagne CD, Ortiz RM, Crocker DE. 2015. Transcriptome analysis of northern elephant seal (Mirounga angustirostris) muscle tissue provides a novel molecular resource and physiological insights. BMC Genomics. 16:64.
- Kim B-M, Ahn D-H, Kang S, Jeong J, Jo E, Kim J-H, Rhee J-S, Park H. 2019. De novo assembly and annotation of the blood transcriptome of the southern elephant seal Mirounga leonina from the south shetland islands, Antarctica. Ocean Science Journal. 54:307–315.
- Latorre D, García-Berthou E, Rubio-Gracia F, Galobart C, Almeida D, Vila-Gispert A. 2020. Captive breeding conditions decrease metabolic rates and alter morphological traits in the endangered spanish toothcarp, aphanius iberus. Int Rev Hydbrobiol. 105:119–130.
- Liu L, Song J, Li J, Huang N, Yang J, Hu S, Ma R, Wang W. 2020. Isoform 1 of fibrinogen alpha chain precursor is a potential biomarker for steroid-induced osteonecrosis of the femoral head. PROTEOMICS – Clinical Applications. 14:1900099.
- Liu Z-f, Ji J-j, Zheng D, Su L, Peng T. 2019. Calpain-2 protects against heat stress-induced cardiomyocyte apoptosis and heart dysfunction by blocking p38 mitogen-activated protein kinase activation. J Cell Physiol. 234:10761–10770.
- Martinez B, Khudyakov J, Rutherford K, Crocker D, Gemmell N, Ortiz R. 2018. Adipose transcriptome analysis provides novel insights into molecular regulation of prolonged fasting in northern elephant seal pups. Physiol Genomics. 50:495–503.
- Moon H-J, Herring S, Zhao L. 2021. Clusterin: a multifaceted protein in the brain. Neural Regen Res. 16:1438.
- Olumuyiwa O, Page M, Soma P, Pretorius E. 2019. Platelets: emerging facilitators of cellular crosstalk in rheumatoid arthritis. Nat Rev Rheumatol. 15:237–248.
- Pan X, Hobbs RP, Coulombe PA. 2013. The expanding significance of keratin intermediate filaments in normal and diseased epithelia. Curr Opin Cell Biol. 25:47–56.
- Patel-Hett S, Wang H, Begonja AJ, Thon JN, Alden EC, Wandersee NJ, An X, Mohandas N, Hartwig JH, Italiano JE. 2011. The spectrin-based membrane skeleton stabilizes mouse megakaryocyte membrane systems and is essential for proplatelet and platelet formation. Blood. 118:1641–1652.
- Patra M, Mukhopadhyay C, Chakrabarti A. 2015. Probing conformational stability and dynamics of erythroid and nonerythroid spectrin: effects of urea and guanidine hydrochloride. PLoS One. 10:e0116991.
- Rosales SM, Thurber RLV. 2016. Brain transcriptomes of harbor seals demonstrate gene expression patterns of animals undergoing a metabolic disease and a viral infection. Peerj. 4:e2819.
- Sadagurski M, Dong XC, Myers MG, White MF. 2014. Irs2 and Irs4 synergize in non-LepRb neurons to control energy balance and glucose homeostasis. Mol Metab. 3:55–63.
- Saeed-Zidane M, Tesfaye D, Shaker Y, Tholen E, Neuhoff C, Rings F, Held-Hölker E, Hoelker M, Schellander K, Salilew Wondim D. 2019. Hyaluronic acid and epidermal growth factor improved the bovine embryo quality by regulating the DNA methylation and expression patterns of the focal adhesion pathway. PLoS One. 14:e0223753.
- Saha K, Hollowell D, Wong PKY. 1994. Mother-to-baby transfer of humoral immunity against retrovirus-induced neurologic disorders and immunodeficiency. Virology. 198:129–137.
- Sano H, Liu SCH, Lane WS, Piletz JE, Lienhard GE. 2002. Insulin receptor substrate 4 associates with the protein IRAS. J Biol Chem. 277:19439–19447.
- Shao Q-Q, Zhang T-P, Zhao W-J, Liu Z-W, You L, Zhou L, Guo J-C, Zhao Y-P. 2016. Filamin A: insights into its exact role in cancers. Pathol Oncol Res. 22:245–252.
- Smith BH, Boesch C. 2011. Mortality and the magnitude of the “wild effect” in chimpanzee tooth emergence. J Hum Evol. 60:34–46.
- Thompson T, Chan Y, Hack A, Brosius M, Rajala M, Lidov H, McNally E, Watkins S, Kunkel L. 2000. Filamin 2 (FLN2): A muscle-specific sarcoglycan interacting protein. J Cell Biol. 148:115–126.
- Toivola DM, Strnad P, Habtezion A, Omary MB. 2010. Intermediate filaments take the heat as stress proteins. Trends Cell Biol. 20:79–91.
- Trukhin A. 2003. Oceanographic and biological conditions affecting the winter distribution of the spotted seal (Phoca largha) in the Sea of okhotsk. Oceanology. 43:387–394.
- Tsuruzoe K, Emkey R, Kriauciunas K, Ueki K, Kahn C. 2001. Insulin receptor substrate 3 (IRS-3) and IRS-4 impair IRS-1- and IRS-2-mediated signaling. Mol Cell Biol. 21:26–38.
- Wang PL, Han JB, Ma ZQ. 2008. Status survey of spotted seal (Phoca largha) in Bohai and Yellow Sea. Chin J Wildlife. 29:29–31.
- Wiśniewski JR, Zougman A, Nagaraj N, Mann M. 2009. Universal sample preparation method for proteome analysis. Nat Methods. 6:359–362.
- Woody S, Zhao L. 2016. Clusterin (APOJ) in Alzheimer’s disease: An Old molecule with a New role. London, United Kingdom: IntechOpen.
- Woulfe J, Checler F, Beaudet A. 1992. Light and electron microscopic localization of the neutral metalloendopeptidase EC 3.4.24.16 in the mesencephalon of the rat. Eur J Neurosci. 4:1309–1319.
- Xiang L, Tzika A, Yingying L, Doninck K, Qian Z, Milinkovitch M. 2010. Preliminary genetic status of the spotted seal Phoca largha in Liaodong Bay (China) based on microsatellite and mitochondrial DNA analyses. Trends Evol Biol. 2:e6.
- Yang L, Xu X, Zhang P, Han J, Li B, Berggren P. 2017. Classification of underwater vocalizations of wild spotted seals (Phoca largha) in Liaodong Bay, China. J Acoust Soc Am. 141:2256–2262.
- Zheng D, Su Z, Zhang Y, Ni R, Fan G-C, Robbins J, Song L-S, Li J, Peng T. 2019. Calpain-2 promotes MKP-1 expression protecting cardiomyocytes in both in vitro and in vivo mouse models of doxorubicin-induced cardiotoxicity. Arch Toxicol. 93:1051–1065.
- Zihlman A, Bolter D, Boesch C. 2007. Skeletal and dental growth and development in chimpanzees of the Taï national park, côte D'Ivoire. J Zool. 273:63–73.